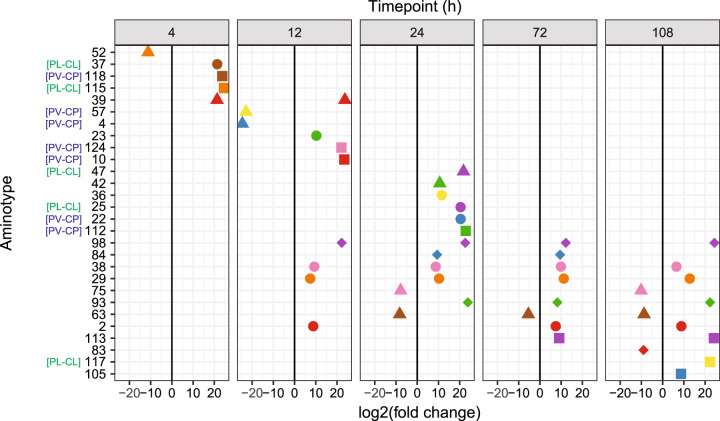
Fig. 4. Differentially abundant Symbiodiniaceae-infecting dinoflagellate RNA virus (“dinoRNAV”) major capsid protein (mcp) “aminotypes” (unique amino acid sequences) across the experiment.
Twenty-eight viral aminotypes (each represented as a point with a unique shape and color) were differentially abundant across control and heated fragments of the coral-symbiont species Pocillopora verrucosa-Cladocopium pacificum (PV-CP) and Pocillopora ligulata-Cladocopium latusorum (PL-CL). Of these, 22 aminotypes occurred at higher relative abundances in heat-treated fragments (right of the 0 lines) and 6 had higher relative abundances in control fragments (left of the 0 lines). Ten aminotypes were differentially abundant at multiple (2–4) timepoints throughout the experiment. This analysis was conducted using DeSeq2 on a non-rarefied dataset including both coral-symbiont species. Aminotypes labeled “[PV-CP]” were unique to PV-CP colonies; aminotypes labeled “[PL-CL]” were unique to PL-CL colonies. DESeq2 analyses were run separately for each time point (t(h) = 4, 12, 24, 72, 108, see Methods for more details).