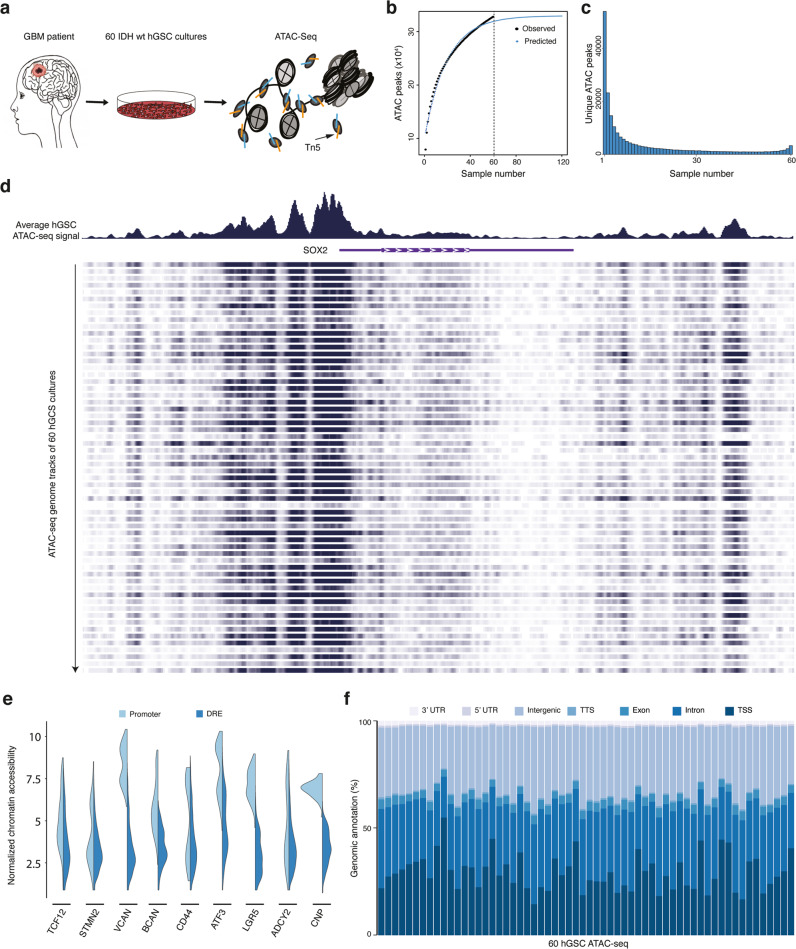
Fig. 2. Heterogeneous chromatin accessibility across 60 patient-derived GSC cultures.
a Overview of 60 patient-derived IDH wild-type GSC cultures analyzed by ATAC-seq. Schematic figures were produced by the authors. b Saturation analysis using a non-linear model. Number of predicted accessible chromatin regions in GBM (blue dotted line), and number of observed accessible chromatin regions in our 60 hGSC samples (black dotted line). c Histogram of the distribution of unique ATAC peaks in the hGSC cohort. d Genome browser tracks of ATAC-seq signals at SOX2. Top panel shows the average genome track of all 60 samples. Bottom panel shows individual results. e Violin plots of cohort-wide distribution of chromatin accessibility at promoters and DRE regions of some GBM meta module and cell lineage-relevant genes. f Genomic annotation of ATAC peaks in each hGSC sample. Source data are provided as a Source Data file for Fig. 2b, c, e, f.