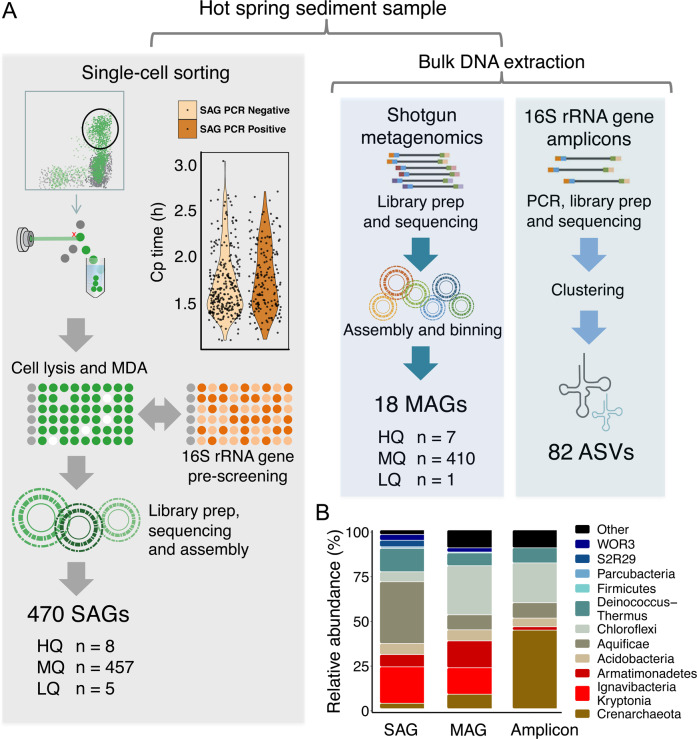
Fig. 1. Production of paired sequence datasets and community composition.
A Workflow for the generation of single amplified genomes (SAGs), metagenome-assembled genomes (MAGs) and 16S rRNA gene amplicons. In the single-cell sorting workflow, the crossing point (cp) value is indirectly proportional to the quantity of amplified DNA as a result of MDA amplification. No statistical difference was observed between 16S rRNA gene PCR positive and negative cp values (p > 0.05), suggesting that the 16S rRNA gene PCR is not a reliable indicator of the total quantity of DNA amplified during whole genome amplification. MAGs were generated from the bulk metagenome based on TNF and coverage profiles of the sediment metagenome, and the 16S rRNA gene amplicons were processed using a standard approach that involved the identification of amplicon sequence variants (ASVs) and classifying the resulting ASVs against the Silva database. B The community composition of this single sediment sample using the three approaches. Specifically, note the lack of Kryptonia and Armatimonadetes in the amplicon data, likely the result of primer mismatch.