Figure 1.
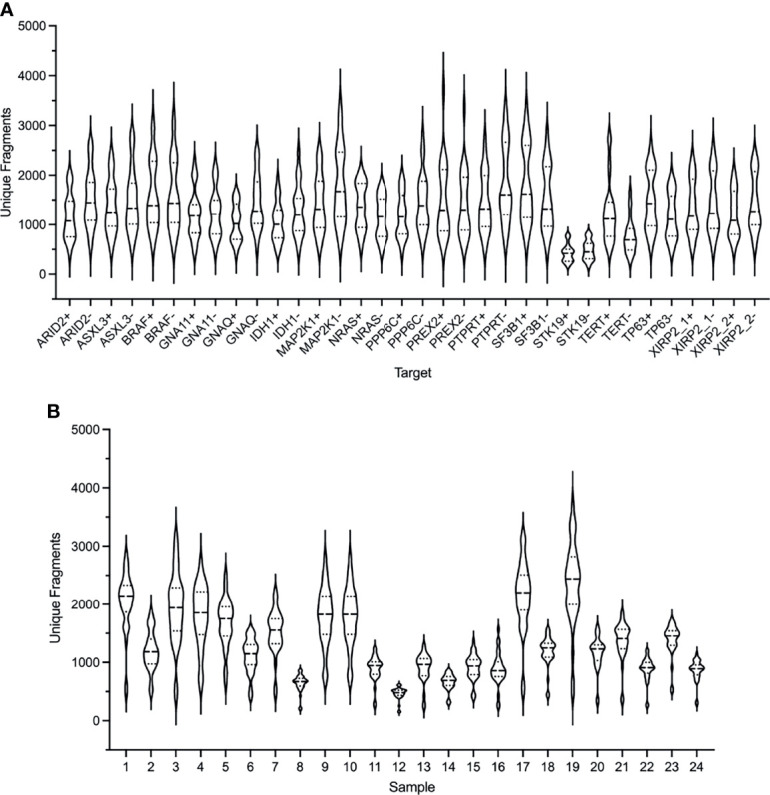
Performance of the ArcherDX custom melanoma ctDNA panel. (A) Distribution of unique fragments for each gene specific target in the custom melanoma panel based on sequencing of 24 samples. For each gene target (see Table S1 for targets) both + and – DNA strand generated fragments are shown. (B) Distribution of unique fragments obtained for each sample based on the 34 targets shown in (A). Samples 1-21 are melanoma patients and 22-24 are healthy controls. Next generation sequencing (NGS) libraries were generated using an ArcherDX Liquidplex NGS workflow followed by Illumina MiSeq sequencing. Unique fragments were defined as deduplicated consensus reads having the same unique molecular barcode. Violin plots show median and SD.
