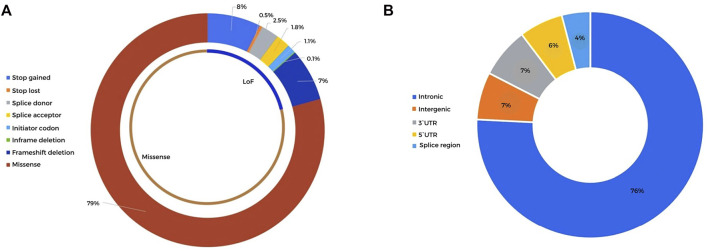
FIGURE 2.
Distribution of the detected high-quality rare coding and noncoding variants detected in 30 HB patients (A). Sequence ontology of the rare coding variants detected after selection by the read depth (>10), Phred score (>20), alternative allele frequency (>0.35), and population frequency (<1%). A total of 2,107 variants were classified into 1,671 missense mutations and 436 LoF variants (B). Sequence ontology of the rare noncoding variants detected after selection by the read depth (>10), Phred score (>20), alternative allele frequency (>0.35), and population frequency (<0.1%). A total of 2,070 noncoding variants were distributed in intronic, intergenic, and 3′ and 5′ UTRs.