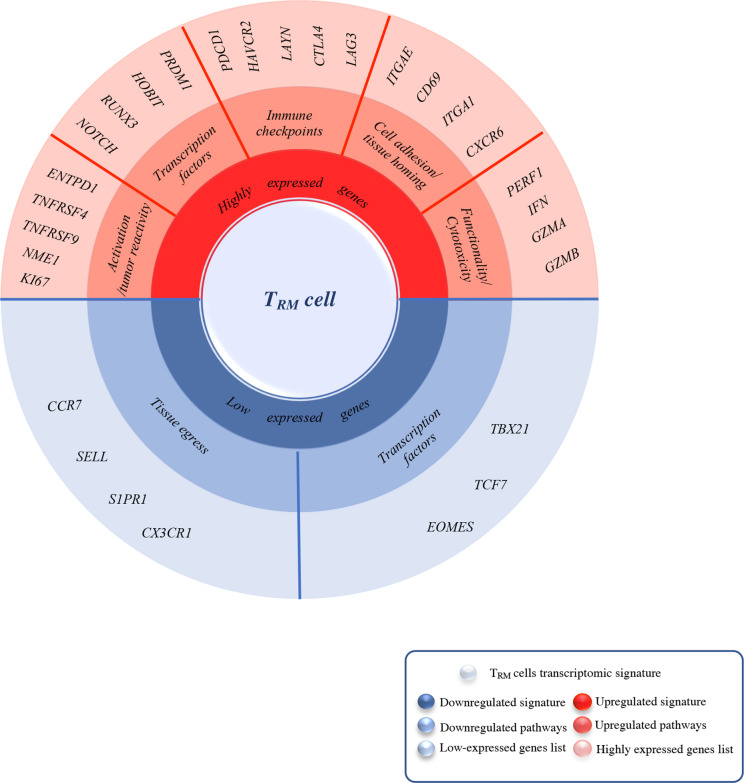
Figure 1.
Summary of the most upregulated/ downregulated genes in TRM cells identified by Bulk/Sc-RNA sequencing. This spin chart summarizes the core residency signature specific to TRM cells compared with TEM and TCM identified by bulk and/or single-cell analysis. TRM signature is mainly characterized by high expression of genes encoding for tissue residency and immune checkpoints (in red), both quite tissue-specific, in this figure, we also focused on those GI cancers specific such as CD103, CXCR6, PD-1, and LAG-3. Another remarkable feature of TRM cells signature is the upregulated expression of cytotoxicity and functionality encoding genes compared with their memory counterparts TEM and TCM. Along with this upregulation, certain genes signaling are either completely shut down (in blue) such as tissue egress ones (SELL, CCR7) and TCF1 which inhibit ITGAE (CD103) expression or downregulated such as TBX21 (T-bet) for IL-15R expression maintenance. GI, gastrointestinal; TCM, central memory T cells; TEM, effector memory T cells; TRM, tissue-resident memory T cells.