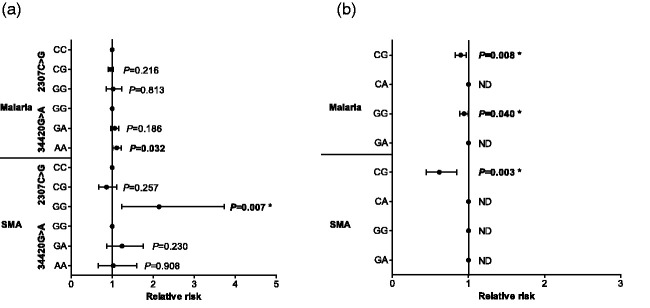
Figure 2.
A Poisson rate regression (R glm function, family=Poisson) was used to explore the relationship between C3 genotypes/haplotypes and the number of malaria and SMA episodes over the three-year follow-up period, with the (logarithm of) age at the patients last visits being the offset variable (rate regression). The Poisson regression was performed with a forward-backward model selection minimizing the AIC. Either C3 genotypes or haplotypes entered the model as covariates. Additional covariates were age at enrollment, sex, HIV, cohort, and sickle cell status (sickle trait and sickle cell disease). Longitudinal relationship between C3 genotypes (2307C>G and 34420G>A), haplotypes (2307C>G/34420G>A), malaria, and SMA. Data presented as relative risk (midline dot) and 95% confidence intervals determined using log-linear regression analyses, adjusting for age at enrollment, sex, HIV, cohort, and sickle cell status (sickle trait and sickle cell disease). Longitudinal follow-up for the cohort was 36 months from the time of enrollment. Correction for multiple comparisons were performed by using the Bonferroni-Holm correction. *significant after Bonferroni-Holm correction. Estimates were not determined (ND) if a covariate was dropped by model selection. (a) Results for models using genotypes as covariates (with the wild types being references absorbed in the models intercept). (b) Results for models with haplotypes as covariates.