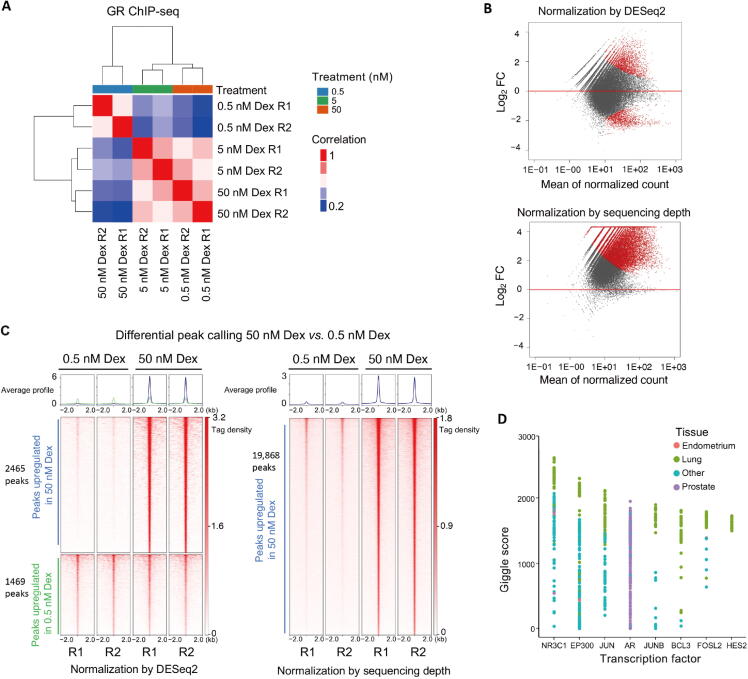
Figure 2.
Example of unsupervised and supervised analyses of differential GR binding in A549 cells. A. Sample–sample heatmap depicting clustering and correlation between A549 cells treated with varying concentrations (0.5 nM, 5 nM, and 50 nM) of Dex in duplicates. B. Visualization of the differences in GR binding between the samples treated with 0.5 nM and 50 nM Dex, plotted using mean of the ChIP-seq peak intensities against Log2 FC of GR binding at the concentrations of 0.5 nM and 50 nM. This illustrates the change in the inferred differential GR binding profile following normalization using scaling factor determined by total reads in peaks (top) and sequencing depth (bottom). C. DeepTools heatmap illustrating differential peaks called by DESeq2 using default scaling factor by total reads in peaks (left) or using scaling factor determined by sequencing depth (right). A group of peaks at the bottom of the left panel exhibit similar binding intensity, however, they are considered downregulated in samples treated with 50 nM Dex, in the peak calling result with default DESeq2 setting. D. Cistrome DB Toolkit analysis result illustrating publicly available ChIP-seq datasets from Cistrome DB ranked by binding profile similarity to gained GR binding sites with Dex treatment. Dex, dexamethasone; GR, glucocorticoid receptor; FC, fold change.