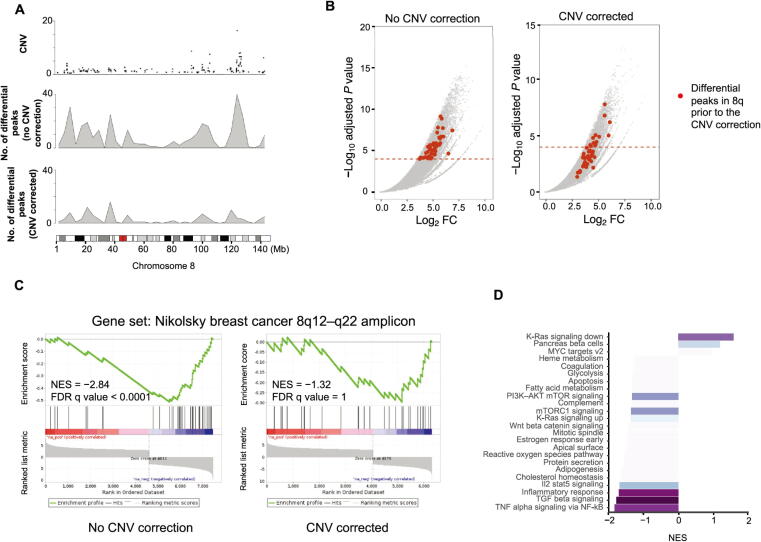
Figure 3.
Identification of differential sites with CNV correction. A. Copy number distribution for an MSS sample on Chr8 (top). Distribution of differentially called peaks without (middle) and with (bottom) CNV correction between MSS and MSI cell lines. The chromosomal plot for Chr8 with genomic coordinate is provided at the very bottom and the red block in the middle indicates the centromere of Chr8. B. Differential peaks in Chr8q called with or without CNV correction. The x axis indicates the Log2 FC of H3K27ac signal difference between MSS and MSI cell lines and y axis indicates −log10 FDR-adjusted P value. Plot on the left shows the peak distribution without CNV correction, and plot on the right shows the peak distribution with CNV correction. Significantly differential (FDR-adjusted P ≤ 0.0001) peaks in Chr8q prior to CNV correction are highlighted in red. Dashed lines indicate the cutoff FDR adjusted P values for significance. C. Enrichment plot for NIKOLSKY_BREAST_CANCER_8Q12_Q22_AMPLICON gene set without (on the left) and with (on the right) CNV correction. D. Enrichment of the GSEA hallmark gene sets after CNV correction based on the differential peak ranking comparing MSS with MSI tumors. MSS, microsatellite stable; MSI, microsatellite instable; CNV, copy number variation; NES, normalized enrichment score; FDR, false discovery rate.