Figure 2. Duplex RT-qPCR detection of SARS-CoV-2 samples.
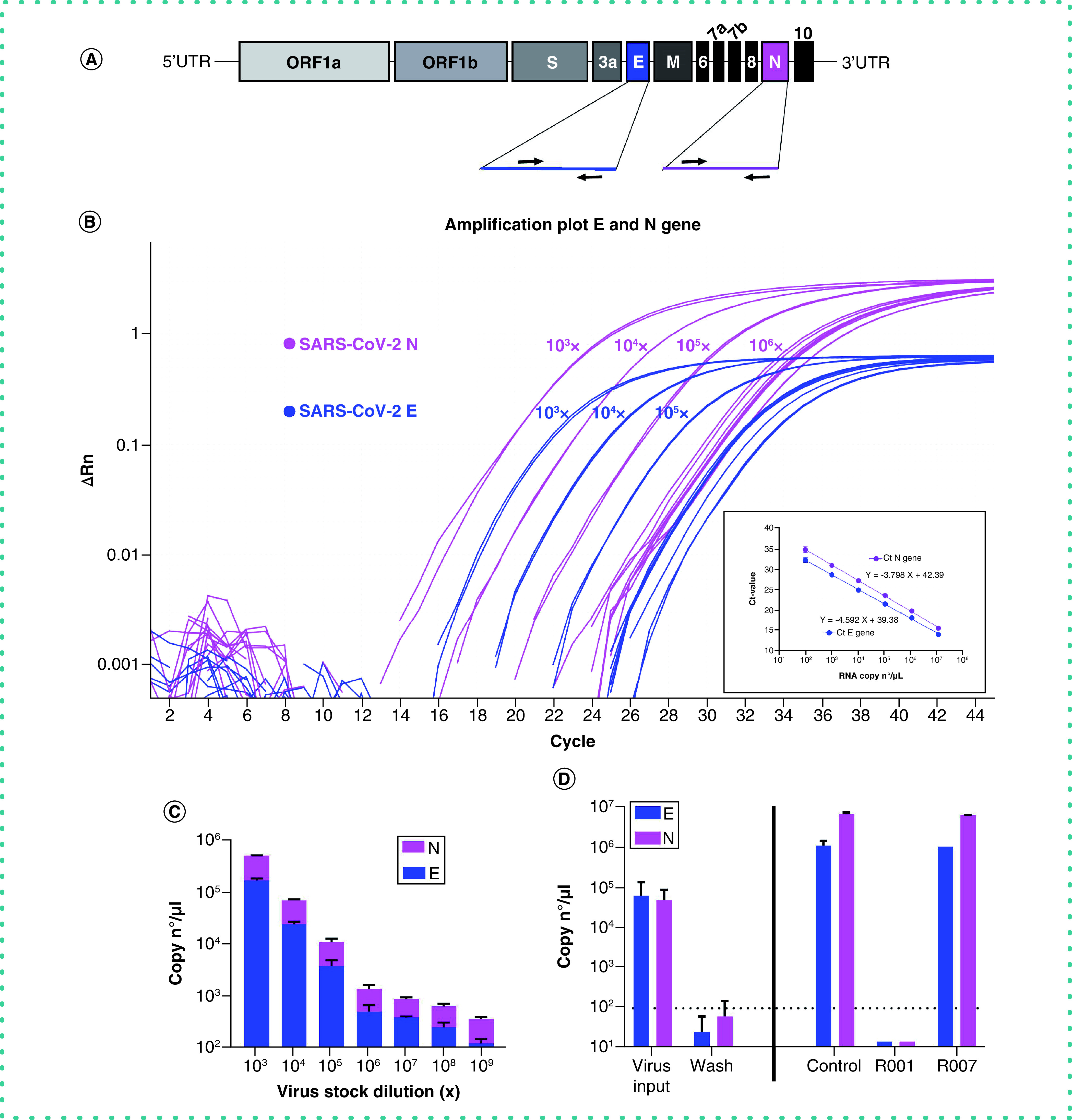
(A) Schematic representation of SARS-CoV-2 genome structure. The full-length RNA genome comprises approximately 30,000 nucleotides and has a replicase complex (comprised of ORF1a and ORF1b) at the 5′UTR. Four genes encode for the structural proteins, i.e., the Spike protein (S), the Envelope protein (E), the Membrane protein (M), and the Nucleocapsid protein (N). The genes for E and N were selected for RT-qPCR detection. (B) Amplification plot of SARS-CoV-2 viral E and N gene. A dilution range (1:10) of a SARS-CoV-2 (20A.EU2 variant) virus stock was measured by duplex RT-qPCR for E and N gene (RNA) copy numbers, starting from a 103x dilution. Graph shows amplification plot for 2 technical replicates of each sample. Insert shows the standard curve for the N and E gene RNA standards. (C) Samples from (B) were analyzed for the quantification of copy numbers of E and N (mean ± standard deviation [SD], n = 2). D) RT-qPCR analysis of the samples from Figure 1. Vero E6 cells were exposed to SARS-CoV-2 (20A.EU2 variant; MOI = 0.015) for 2 h. Supernatant (= virus input) was then removed, and cells were washed in PBS (= wash) and given fresh medium. In the conditions with the antibody-treatment (R001 or R007), 2.5 μg/ml of antibody was administered together with the virus, and administered again after the wash step. At day 3 postinfection, supernatant was collected and a duplex RT-qPCR was performed to quantify the copy numbers of E and N. The dotted line refers to the residual amount (background) of virus that attached aspecifically to the cells. For R001 a complete protection of virus entry was obtained, whereas R007 had no antiviral effect. For each sample, the average (mean ± SD) of two technical replicates is given (n = 2), plotted on a log10 scale.
