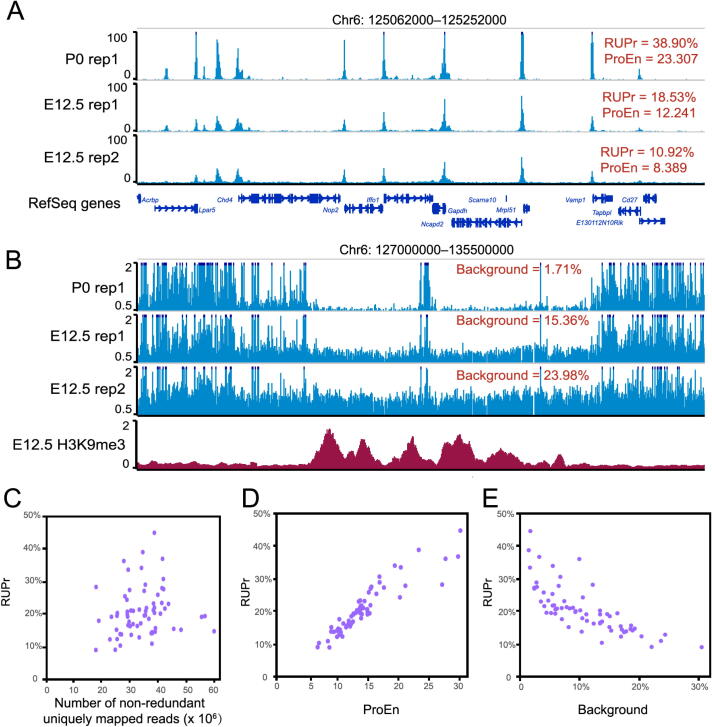
Figure 2.
Key QC metrics of ATAC-seq data A. Forebrain ATAC-seq datasets (sequencing depth normalized to 10 million reads) with distinct RUPr and ProEn (top to bottom: high to low) were visualized on the WashU Epigenome Browser. B. Forebrain ATAC-seq datasets (same as A) with distinct background values (top to bottom: low to high) in the heterochromatin regions highly enriched with repressive H3K9me3 histone modification in the mouse forebrain. C. Relationship between the number of non-redundant uniquely mapped reads and RUPr in ENOCDE mouse ATAC-seq datasets. D. Relationship between ProEn and RUPr in ENOCDE mouse ATAC-seq datasets. E. Relationship between background and RUPr in ENOCDE mouse ATAC-seq datasets. QC, quality control; RUPr, reads under peak ratio; ProEn, promoter enrichment; E12.5, embryonic day 12.5; P0, postnatal day 0.