Figure 3.
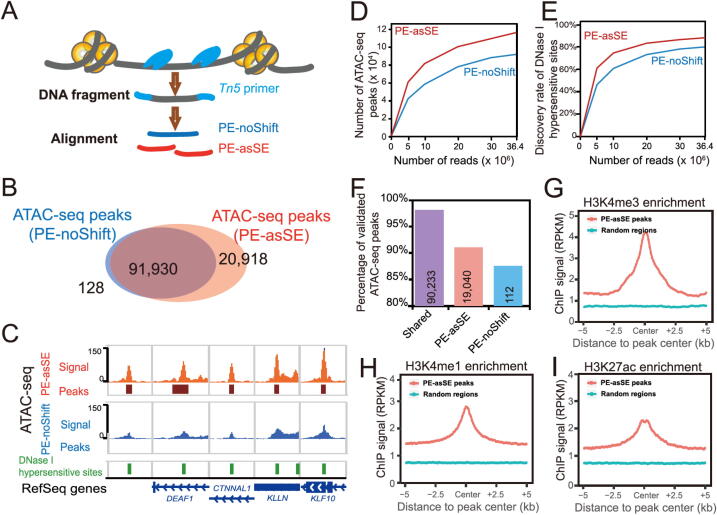
AIAP data processing strategy (PE-asSE) performs better than classic method (PE-noShift) in OCR identification A. Schematic representation of PE-noShift and PE-asSE data processing strategy. PE-noShift representing the one complete DNA fragment insertion was generated as in previous studies [26], [27], [28], [29], [30], [31], [32]. PE-asSE representing two DNA fragment insertions was generated by AIAP. B. Venn diagram showing ATAC-seq peaks identified with PE-noShift and PE-asSE in the GM12878 cell line. C. ATAC-seq data of GM12878 cell line were independently processed as PE-asSE (red) and PE-noShift (blue), and visualized together with DHSs of GM12878 cell line (green) on the WashU Epigenome Browser. Five regions were randomly selected that contain the peaks identified only in PE-asSE mode but not in PE-noShift mode. D. Number of ATAC-seq peaks identified with PE-noShift mode and PE-asSE mode at different sequencing depths by randomly sampling the Omni-ATAC-seq data of GM12878 cell line [31]. E. Discovery rate of DHSs of GM12878 cell line by PE-asSE and PE-noShift at different sequencing depths (randomly sampling same as in D). F. Percentage of shared, PE-asSE-specific, and PE-noShift-specific ATAC-seq peaks validated by DHSs of GM12878 cell line identified by ENCODE. PE-asSE-specific ATAC-seq peaks were highly enriched with active histone modifications H3K4me3 (G), H3K4me1 (H), and H3K27ac (I), when compared to 20,000 regions randomly selected from the genome (green). DHS, DNase I hypersensitive site.