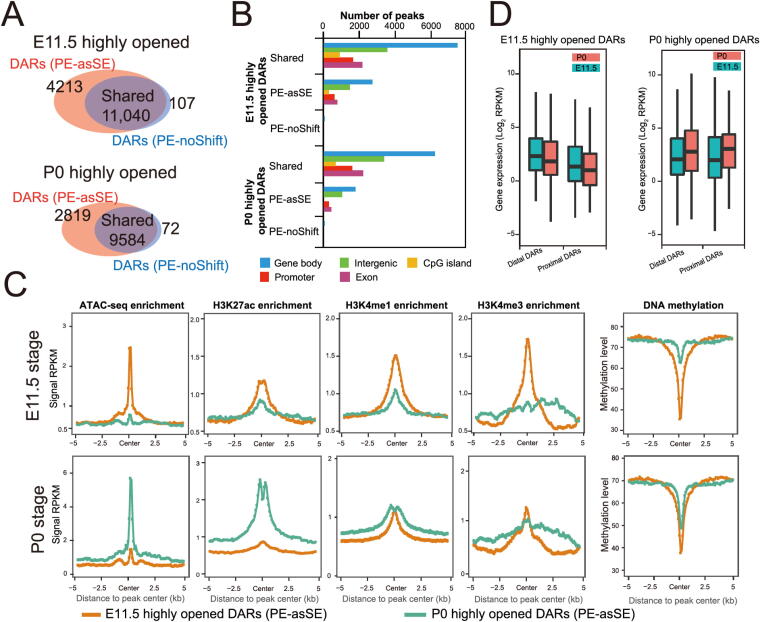
Figure 4.
AIAP data processing strategy (PE-asSE) performs better than the classic method (PE-noShift) in DAR identification A. Venn diagram showing DARs identified with PE-noShift mode and PE-asSE mode when comparing ATAC-seq data collected from mouse liver at E11.5 and those from P0. B. Genomic distribution of 20,624 shared, 7032 PE-asSE-specific, and 179 PE-noShift-specific DARs. C. Enriched epigenetic modifications (left to right: ATAC-seq, H3K27ac ChIP-seq, H3K4me1 ChIP-seq, H3K4me3 ChIP-seq, and DNA methylation) on the PE-asSE-specific DARs from mouse liver at E11.5 (top) and P0 (bottom). D. The expression of genes associated with PE-asSE-specific E11.5 DARs (left) and P0 DARs (right) in proximal (2 kb around TSSs) and distal (2–20 kb around TSSs) at the two developmental stages. TSS, transcription start site.