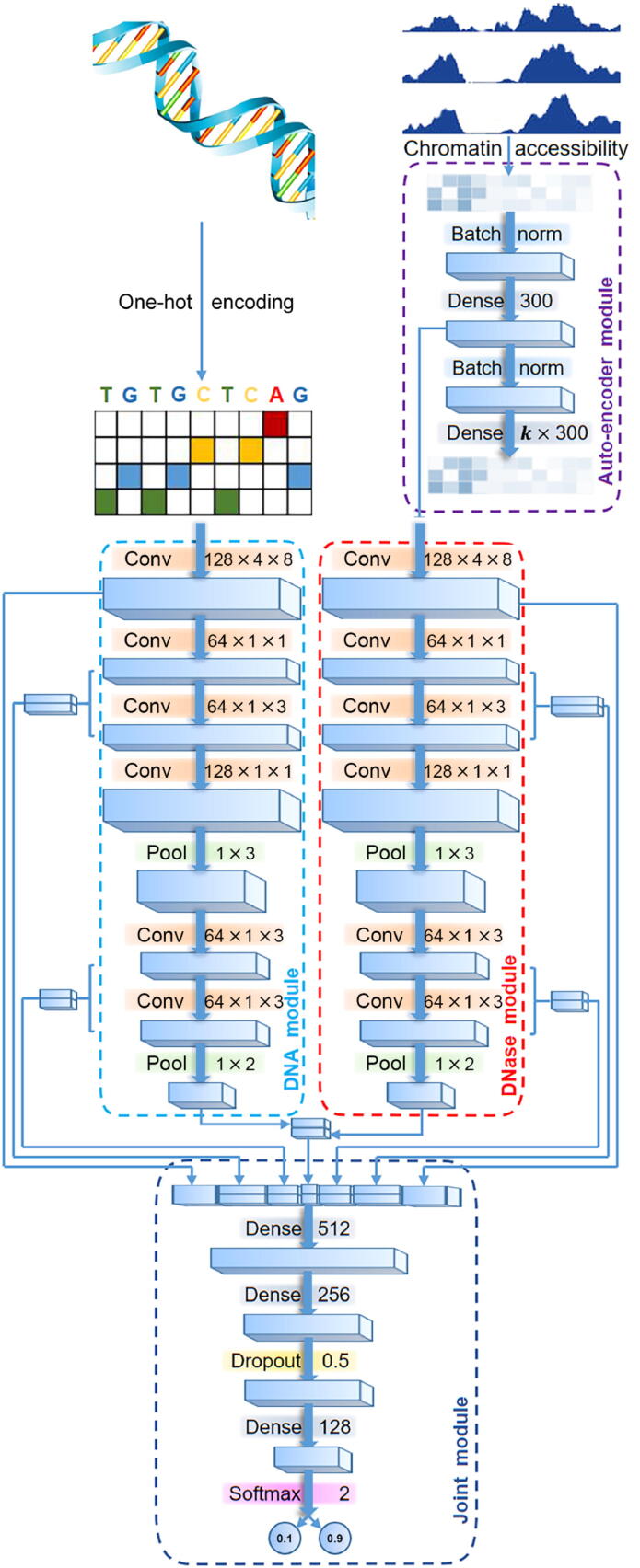
Fig. 1.
Graphical illustration of DeepCAPE. First, a DNA module is used to extract features of the input DNA fragment. Second, an auto-encoder module is adopted to embed DNase-seq data into a low-dimensional space. Third, a DNase module is used to extract features of chromatin accessibility after dimensionality reduction. Finally, a joint module integrates outputs of the DNA and DNase modules to predict the probability that an input sequence is an enhancer. Conv, convolutional layer; Pool, max-pooling layer; Batch norm, batch-normalization layer.