Abstract
Understanding the joint roles of protein sequence variation and differential expression during adaptive evolution is a fundamental, yet largely unrealized goal of evolutionary biology. Here, we use phylogenetic path analysis to analyze a comprehensive venom-gland transcriptome dataset spanning three genera of pitvipers to identify the functional genetic basis of a key adaptation (venom complexity) linked to diet breadth (DB). The analysis of gene-family-specific patterns reveals that, for genes encoding two of the most important venom proteins (snake venom metalloproteases and snake venom serine proteases), there are direct, positive relationships between sequence diversity (SD), expression diversity (ED), and increased DB. Further analysis of gene-family diversification for these proteins showed no constraint on how individual lineages achieved toxin gene SD in terms of the patterns of paralog diversification. In contrast, another major venom protein family (PLA2s) showed no relationship between venom molecular diversity and DB. Additional analyses suggest that other molecular mechanisms—such as higher absolute levels of expression—are responsible for diet adaptation involving these venom proteins. Broadly, our findings argue that functional diversity generated through sequence and expression variations jointly determine adaptation in the key components of pitviper venoms, which mediate complex molecular interactions between the snakes and their prey.
Keywords: genotype–phenotype, venom, diversity, adaptation, diet breadth
Introduction
Adaptation at the molecular level can occur through changes in protein-coding sequence or the patterns of gene expression, and identifying the relative roles of these mechanisms is central to understanding trait evolution (Barrett and Hoekstra 2011; Rockman 2012; Rausher and Delph 2015; Smith et al. 2020). Although both mechanisms play important roles in evolution (Carroll 2005, 2008; Hoekstra and Coyne 2007), there are differing expectations for their relative contributions to complex traits. Protein-coding mutations can produce novel functions, especially when coupled with gene duplications that reduce selective constraints (Ohno 1970; Hoekstra and Coyne 2007). Regulatory changes serve critical roles in morphological evolution, and the time and tissue-specific nature of gene expression is expected to reduce the pleiotropic effects of regulatory variation, facilitating the evolution of novel adaptations (Carroll 2008; Stern and Orgogozo 2008). Moreover, because there are more pathways for altering the expression of a gene compared with altering its sequence, regulatory mechanisms present larger mutational targets, which lead to differences in their evolutionary rates and lability compared with protein-coding regions (Rokyta, Wray et al. 2015; Besnard et al. 2020). Understanding how protein-coding and/or regulatory changes mediate realized adaptive function has significant implications for identifying general evolutionary processes linking genomic variation to adaptive phenotypes (Smith et al. 2020). This requires the development and use of detailed genotype–phenotype maps that are linked to realized ecological variation from diverse species groups.
Traditionally, genotype-to-phenotype maps for adaptive traits have been constructed using a “forward genetics” approach which focuses on experimental analyses of segregating genetic variation in model species (Barrett and Hoekstra 2011). Forward genetics has proved highly successful for identifying the molecular basis of many adaptations, but is limited by the need to work with model species amenable to either experimental manipulation or observational studies that link segregating genetic variants to phenotypes with statistical association methods (Tanksley 1993; Marigorta et al. 2018). These methods are incompatible with many adaptive phenotypes of interest to evolutionary biologists because such traits may occur in species that cannot be interbred or where the phenotypic variation of interest may only occur between species (Smith et al. 2020). Studies to date are limited to a small number of species in which the “forward genetics” paradigm can be applied, which raises questions about the generality of their results, especially at macroevolutionary scales.
A recently proposed approach to overcome these issues is to use comparative phylogenetic methods to analyze clade-wide genomic datasets to link phenotypic variation to its genetic underpinnings (Nagy et al. 2020; Smith et al. 2020). This approach builds on the increasing availability of genomic datasets and uses the long-standing comparative phylogenetic methods to identify associations between functionally relevant genetic and phenotypic variation while accounting for a shared ancestry (Smith et al. 2020). Although lacking the experimental certainty of forward genetic approaches, comparative phylogenetics methods broaden the scope of studies of adaptive phenotypes and can yield new insights into how evolutionary mechanisms mold the genetic basis of phenotypic variation (Pease et al. 2016; Hu et al. 2019; Sackton et al. 2019). Comparative methods like phylogenetic path analysis that test for a causal structure among a suite of compared variables have recently been used to understand genome–environment interactions in multiple groups (von Hardenberg and Gonzalez-Voyer 2013; Voyer and Garamszegi 2014; Guignard et al. 2019; Chak et al. 2021). Phylogenetic path analysis, therefore, provides a useful method to apply to genome scale data for analyzing functional genetic variation from multiple species, especially when the genetic and phenotypic variations are closely tied to ecological functions.
Animal venoms are a model system for investigating the molecular mechanisms that underlie adaptive traits because of the unusually direct connection between venom genes, phenotypes, and adaptive function that allows comprehensive investigation across multiple levels of biological organization (Gibbs and Rossiter 2008; Casewell et al. 2012, 2014; Rokyta, Margres, et al. 2015). Whole venoms are complex adaptive phenotypes that can be broken down into distinct components—individual proteins making up the venom—and linked to known molecular underpinnings, and their functional impacts (Casewell et al. 2013; Zancolli and Casewell 2020). Several of the major gene families that contribute to venom occur as tandemly arrayed gene islands in distinct genomic locations (Sanggaard et al. 2014; Gendreau et al. 2017; Casewell et al. 2019; Schield et al. 2019; Margres et al. 2021). This genomic architecture means the evolution of venom genes and the pathway from genotype to a complex phenotype can be investigated in multiple gene families across a set of venomous species. These features make venom an exceptional system for examining how complex adaptive phenotypes are assembled and evolve, and for understanding the impact of phenotypic complexity on ecological function (Holding, Drabeck, et al. 2016; Sunagar et al. 2016; Arbuckle 2020; Giorgianni et al. 2020; Zancolli and Casewell 2020; Holding et al. 2021).
Studies of venomous species have yielded numerous important insights into how molecular adaptations arise. For example, molecular and ecological studies in cone snails have provided evidence for the dynamic expansion of toxin gene families, evidence of pervasive positive selection, and correlations between venom compositions and diet (Duda and Palumbi 1999, 2004; Duda and Remigio 2008; Remigio and Duda 2008; Chang and Duda 2012, 2014; Phuong et al. 2016; Li et al. 2017). In spiders, venom complexity has been shown to vary based on feeding ecologies (Pekár et al. 2018). Several studies on individual snake species have also evaluated the roles of sequence and expression evolution in venom toxins and indicate that both mechanisms facilitate phenotypic evolution, possibly in different evolutionary or ecological contexts (Margres et al. 2016; Margres, Bigelow et al. 2017; Margres, Wray et al. 2017; Hofmann et al. 2018; Rautsaw et al. 2019; Zancolli et al. 2019).
At the macroevolutionary scale, a recent study by Holding et al. (2021) used k-mer based metrics from venom-gland transcriptomes and whole venom RP-HPLC data from 68 primarily North American pitvipers (rattlesnakes and moccasins) to show a strong positive relationship between the molecular complexity of venom and phylogenetic diversity in diet. This study identified the molecular complexity of venom as an adaptive phenotype that is correlated with a key ecological trait (diet breadth [DB]) in these snakes, although their reliance on k-mers prevented the specific genetic mechanisms from being identified. Nonetheless, the availability of a comprehensive molecular dataset on venom variation for a phylogenetically diverse snake clade opens the door to using a comparative phylogenetics approach to identify the specific genetic mechanisms underlying this adaptive trait.
Here, we analyze fully assembled venom-gland transcriptomes for the 68 lineages represented in Holding et al. (2021) using the phylogenetic path analysis (von Hardenberg and Gonzalez-Voyer 2013; Voyer and Garamszegi 2014) to dissect the relative roles of gene composition, protein sequence diversity (SD), and expression diversity (ED) as they relate to DB in these snakes. In addition, we capitalized on the nature of venom as a mixture of proteins from distinct multi-gene families to determine if separate or concerted evolutionary processes contribute to venom diversity from separate regions of the genome. Finally, for two families where toxin SD showed significant associations with dietary breadth, we tested whether lineages show evidence for similar or divergent evolutionary pathways for generating protein SD. Our results show that both SD and expression variation mediate adaptation in pitviper venoms, but the roles of SD and expression vary for different components of this complex phenotype. These results highlight how complex molecular traits can evolve via alternative routes to adaptation.
Results
Venom-Gland Transcriptomes
We assembled and annotated venom-gland transcriptomes for the 214 individuals comprising 68 rattlesnake and moccasin lineages used in Holding et al. (2021), with specimen representation for each lineage varying from 1 to 10 individuals (supplementary tables S1 and S2, Supplementary Material online). Individual snakes expressed on average 78.4 transcripts encoding toxin proteins (range = 32–128). Using the annotated transcriptomes, we calculated gene content (GC) as the total number of toxins, toxin SD as the effective number of amino acid 20-mers (the number of unique k-mers that would represent equivalent diversity with uniform occurrence, see Materials and Methods), and toxin ED as the effective number of expressed toxin transcripts (the number of expressed toxins that would represent equivalent diversity with uniform expression, see Materials and Methods). Lineage-specific estimates of these measures were obtained by averaging across samples, though variation in these metrics was apparent within several lineages (supplementary figs. S1–S3, Supplementary Material online).
To verify that technical variation in sample treatment (e.g., differences in sequencing the depth and numbers of assembled transcripts) did not bias statistical inference, we tested for a relationship between these variables and the number of recovered toxins. Although we found some evidence of a marginally significant correlation between the number of recovered toxins and the number of merged reads among samples (P = 0.063, supplementary fig. S4, Supplementary Material online), this relationship explained a relatively small amount of variation (R2 = 0.016). Similarly, we found no significant relationship between the number of expressed transcripts and recovered toxins (P = 0.664, R2 < 0.001, supplementary fig. S5, Supplementary Material online). Importantly, we found no evidence of an interaction between the number of merged reads (P = 0.369, supplementary table S3, Supplementary Material online) or expressed transcripts with lineage assignment (P = 0.618, supplementary table S4, Supplementary Material online), indicating that inferences made among lineages are unbiased by technical variation.
We tested for evidence of phylogenetic signal among GC, SD, and ED metrics with Blomberg’s K and lambda. GC, SD, and toxin ED all showed evidence of significant phylogenetic signal based on estimates of Blomberg’s K (GC = 0.47, ED = 0.38, SD = 0.46), and both GC and SD showed evidence of significant phylogenetic signal based on lambda (supplementary table S5 and fig. S6, Supplementary Material online). Evidence of phylogenetic signal in these metrics indicates a moderate degree of predictability in the venom genotype-to-phenotype map based on the degrees of evolutionary divergence among related snake lineages.
Path Analysis
To examine how expression and protein-coding sequence evolution affect the dynamics of venom and diet diversity, we tested 10 path models defining hypothesized relationships among GC, SD, ED, and DB (supplementary fig. S7, Supplementary Material online) for 30 snake lineages, for which we had reliable diet data. Here, DB corresponded to the mean phylogenetic distance (MPD) measure of diet used in Holding et al. (2021), who showed that snake DB as a function of its phylogenetic diversity of prey species was a better predictor of venom complexity than prey species richness alone. Phylogenetic path models represented varying roles of SD and ED as having direct or indirect effects on DB, independently or in combination, whereas GC was modeled as acting indirectly through these variables.
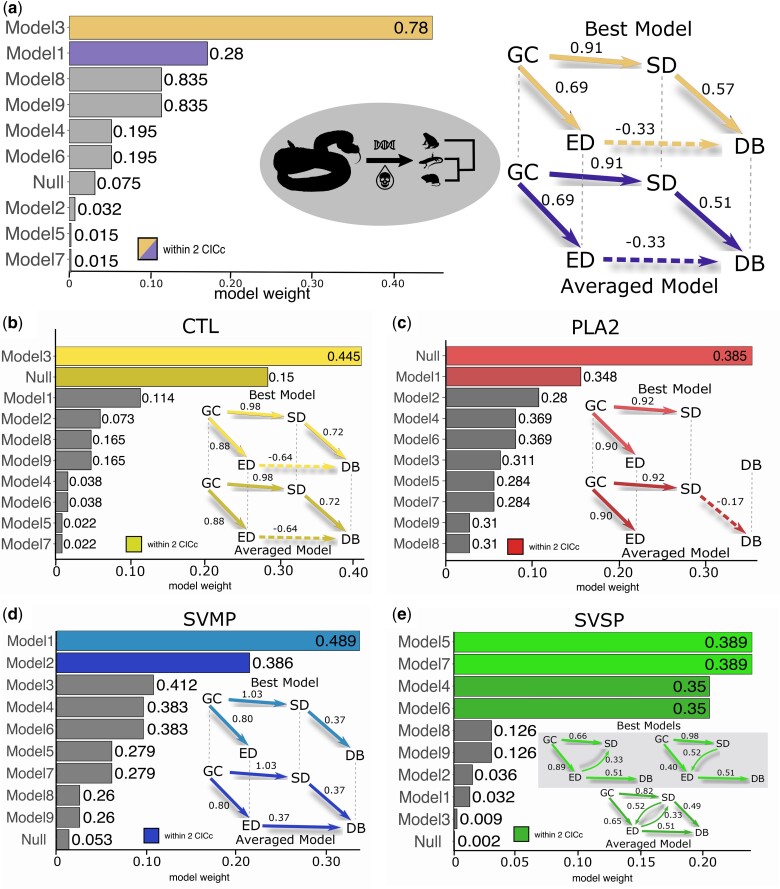
We found the highest support for Model 3 in which SD had a moderate, positive correlation with DB, and surprisingly, ED had a moderate negative correlation with DB (fig. 1, supplementary fig. S8 and table S6, Supplementary Material online). Hence, snakes with more diverse, but less evenly expressed sequences had broader diets. As expected, GC was positively correlated with SD and ED in this model, showing a strong indirect association with DB mediated through SD and expression. However, support for Model 3 was not absolute. Model 1 was within the 2 C statistic Information Criterion (CICc) of Model 3, indicating similar statistical support (fig. 1, supplementary fig. S3, Supplementary Material online). Unlike Model 3, Model 1 did not include a connection between ED and DB, and showed a weaker relative relationship between SD and diet (supplementary fig. S8, Supplementary Material online). Because of the overall similarity of Model 3 and Model 1, the weighted average model we recovered was similar to Model 3 (fig. 1).
Fig. 1.
Path analysis for models of venom evolution and DB for an overall venom model (a) and the CTL (b), PLA2 (c), SVMP (d), and SVSP (e) toxin families. Path models test for varying effects among GC, SD, ED, and DB and are defined in supplementary figure S7, Supplementary Material online. The barplots show model weights for CICc comparisons. Numbers adjacent to the bars represent p-values for the test of the null hypothesis that the model fits the data structure. Models with P < 0.05 are statistically untenable. The best performing and averaged models are shown at the right with path coefficients (partial regression coefficients standardized to the other independent variables) indicated by numbers adjacent to arrows. Dashed lines in the graphical models indicate negative relationships. Averaged models were calculated based on a model weight of all top models within two CICc.
In both top-performing models, SD and ED predicted changes in diet. Importantly, although our path models modeled venom SD and ED as predictors of DB, these relationships do not imply directional causality. Rather, the direct positive correlation between SD and DB indicates that increased sequence variation is associated with more diverse diets. Sequence variation, in turn, is heavily influenced by the underlying GC. In contrast, a more even, and hence, diverse toxin expression is associated with a narrower diet. Next, we sought to explore this initially counterintuitive result for ED in more detail.
We suspected that the analysis of pooled data may obscure more subtle relationships between expression and DB for individual toxin gene families which, because they are found at distinct genomic locations in these snakes (Schield et al. 2019), represent semi-independent replicates of how venom complexity evolves. To examine whether the patterns of complexity detected for the whole venom phenotype are representative of the patterns found in individual toxin families, we tested the possible path models in four tandemly arrayed toxin families: C-type lectins (CTLs), phospholipase A2s (PLA2s), snake venom metalloproteases (SVMPs), and snake venom serine proteases (SVSPs). These toxin families have previously shown heterogeneous relationships between expressed transcript sequence complexity (measured in k-mers) and DB, with three of the families having positive relationships, whereas CTLs displayed no relationship (Holding et al. 2021).
Here, we report substantial differences in the optimal models for family-specific path analyses. In particular, the analyses of SVMP and SVSP families separately showed support for models where both SD and ED had direct positive correlations with DB (fig. 1d and e). Thus, in contrast with the overall analyses, within each of these toxin families, more diverse patterns of expression were associated with increased DB. All competitive models for the SVSP family also supported a direct relationship between SD and ED. Models with opposing directions of the relationship between SD and expression showed equivalent support, as expected, but varied in effect estimates (fig. 1e). This finding indicates an interacting effect of the sequence and expression evolution in SVSPs where increased SD and more even toxin expression are linked.
In contrast, for analyses of the CTL and PLA2 gene families, the top ranked model set included the null model, which did not include any direct connection between sequence and ED and DB (fig. 1b and c). This result suggests that ‘functional diversity’ in CTLs and PLA2s does not influence the ability of these snakes to consume phylogenetically diverse prey but that other characteristics, such as the total expression or the presence of paralogs with specific functions, may play more important roles for these toxin family.
Variation in Expression
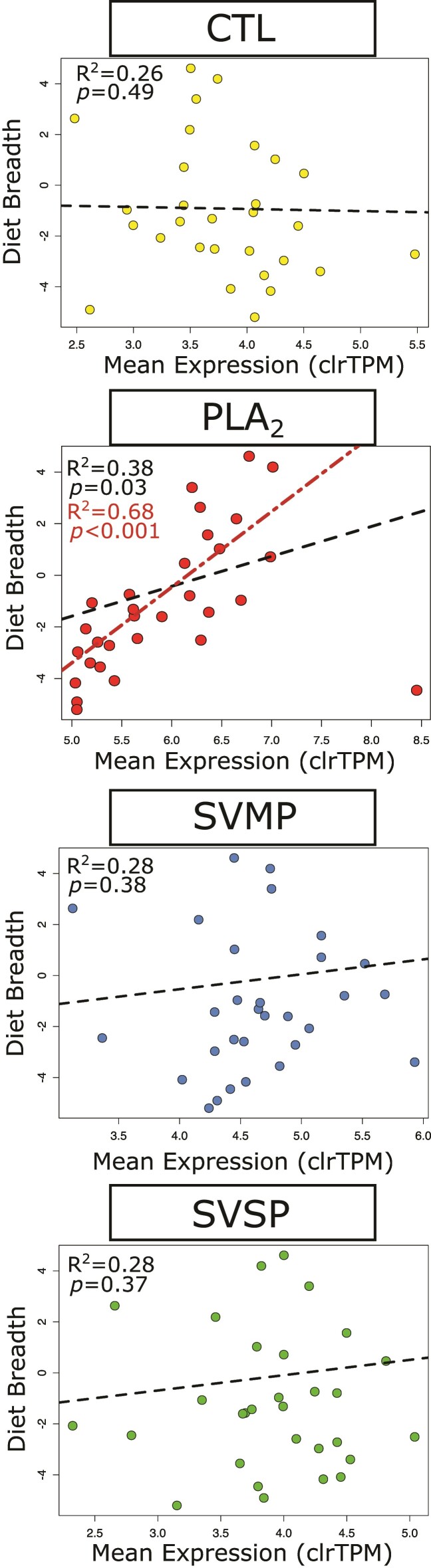
To explore how other aspects of venom composition are associated with DB, we compared how absolute expression patterns (rather than complexity in expression) varied among and within major families, and tested for correlations with DB. As expected, the number and mean expression of toxins varied significantly among families with PLA2s exhibiting the lowest number of toxins per lineage (P < 0.001, supplementary fig. S9 and table S2, Supplementary Material online), but the highest mean expression levels (P < 0.001, supplementary fig. S9, Supplementary Material online). PLA2s also exhibited a positive correlation between mean expression and DB (P = 0.03, R2 = 0.38) (fig. 2). This relationship becomes even stronger when a single, high leverage outlier (the South American Rattlesnake, Crotalus durissus) is excluded from the analysis (P < 0.001, R2 = 0.68; fig. 2).
Fig. 2.
Comparison of DB and mean expression for CTLs, PLA2s, SVMPs, and SVSPs. Mean expression is measured as center log-ratio transformed TPM. Black dashed lines indicate the lines of best fit inferred with phylogenetic linear models. The red dotted and dashed line and lower R2 and P-value in PLA2s displays the line of best fit if the outlying datapoint for C. durissus is excluded.
These relationships explain why the global path analysis shows a negative relationship between ED and DB. The indices used for path analyses measure diversity as a function of richness and relative abundance. ED specifically is derived from the number of expressed transcripts and their relative expression (evenness), where we consider more even expressions to be more complex. Because PLA2s consist of only a few, often highly expressed transcripts, they exert a disproportionate effect on expression evenness. Thus, lineages with more complex diets with more highly expressed PLA2s can show less diverse expression patterns overall. In sum, the strong positive relationship between the mean PLA2 expression and DB suggests that abundance rather than compositional diversity of PLA2s facilitates eating a broader range of prey.
Mechanisms of Gene-Family Diversification
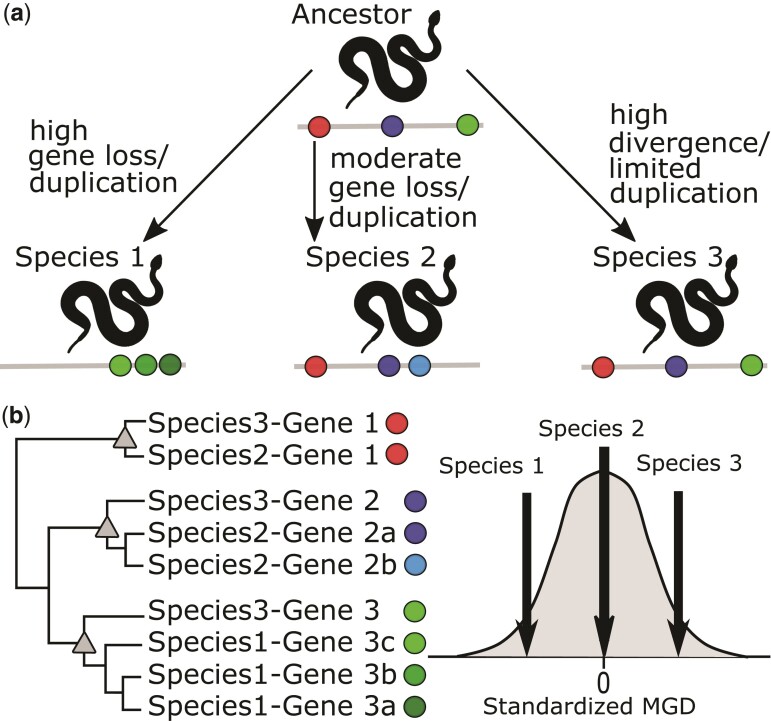
Our analysis showed that the SVMP and SVSP venom gene families both showed evidence of positive relationships between amino acid SD and DB. In large gene families, gene SD is inextricably linked to gene duplications and divergence which collectively produce diverse paralogs. Most pitviper lineages express multiple SVMP and SVSP toxin paralogs and the diversity of these toxin assemblages can lend insight into the patterns of gene diversification. Ancient duplications may be observed as highly divergent paralogs in modern taxa, but recent duplications also occur in many venom gene families (Wong and Belov 2012; Giorgianni et al. 2020). The assemblage of toxin paralogs in the venom of a given lineage may consist primarily of conserved ancient paralogs, less divergent recent paralogs, or a combination (fig. 3a). Each of these scenarios can generate sequence variation, but whether either is overrepresented as an evolutionary pathway in venoms is not clear.
Fig. 3.
Graphical representation of how MGD informs the understanding of the patterns of gene-family diversification. (a) Three hypothetical lineages descending from a common ancestor with differing patterns of gene diversification. Individual genes are shown as colored circles on gray lines. (b) Hypothetical gene-family phylogeny derived from the three lineages in (a) and a representation of hypothetical MGD metrics based on the phylogeny.
To assess what patterns of paralog diversification characterized venom gene diversity, we used a similar method to that of Chang and Duda (2014) to compare the within-family toxin diversity of each individual against the within-family toxin diversity across Agkistrodon, Crotalus, and Sistrurus. Specifically, we calculated phylogenetically weighted, standardized mean genetic distance (MGD) for two toxin families where we expected paralog diversification could have an ecological impact acting through SD: SVMPs and SVSPs. The standardized values of MGD represent the diversity of toxins in a toxin family (i.e., SVMPs or SVSPs) expressed by an individual compared with the total diversity of the toxin family. In the context of a gene family, low estimates of assemblage MGD would occur through the assemblages of highly similar (phylogenetically clustered) paralogs, whereas high estimates of MGD would result from assemblages that were very diverse (phylogenetically dispersed) (fig. 3b). This approach, therefore, allowed us to infer whether diversity in these families arose primarily through expression/reliance on highly divergent genes such as ancient or highly derived paralogs versus clusters of more recently duplicated, less differentiated paralogs (fig. 3b).
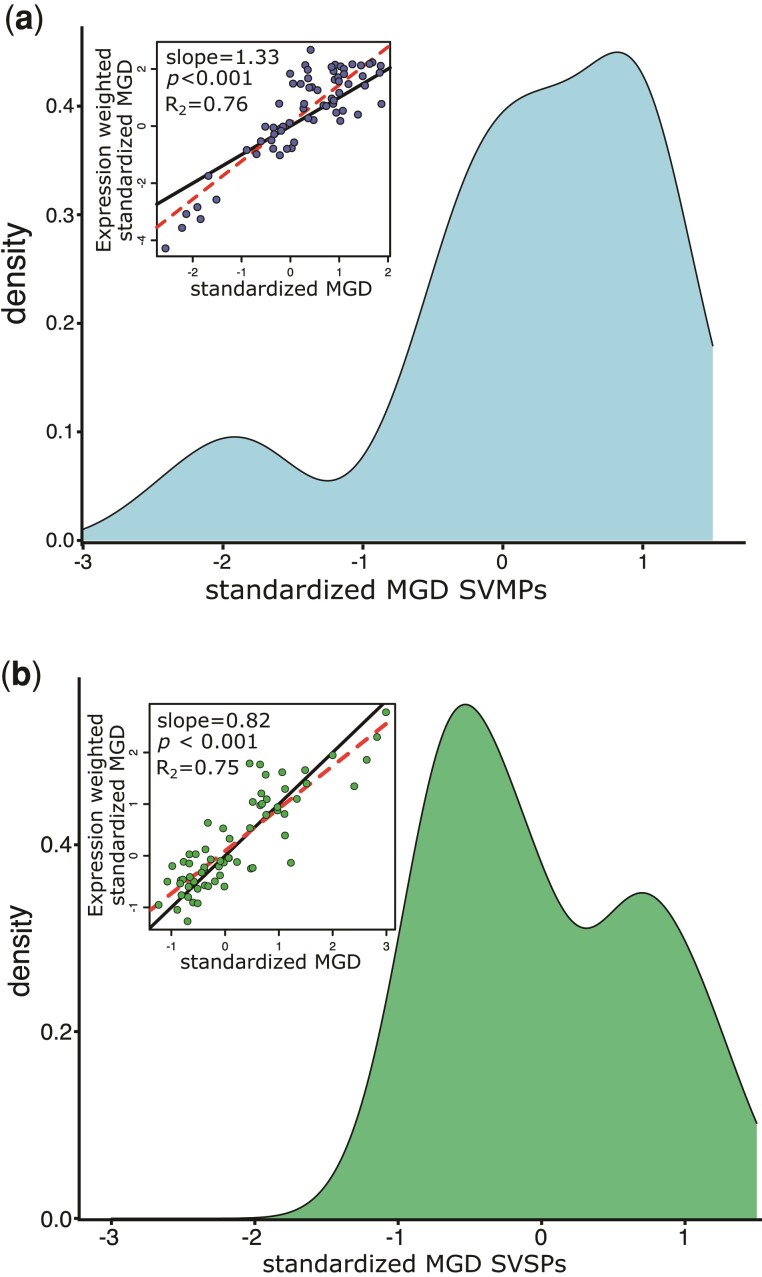
We observed a range of negative and positive standardized MGD values for SVMPs and SVSPs, with slightly positive means for the overall distribution for both families (mean SVMP = 0.29, median SVMP = 0.39, mean SVSP = 0.21, median SVSP = −0.03, supplementary figs. S10 and S11, Supplementary Material online). These results indicate that on average, expressed genes tend to be more divergent than would be expected by chance alone. However, both the SVMP and SVSP distributions appeared multimodal (fig. 4) and Wilcoxon signed rank tests found the distribution of SVMP standardized MGD values to be different than 0 (P = 0.005), although SVSPs were not (P = 0.247). In the case of SVMPs, two clear peaks were visibly centered at approximately −2 and 0.5, with some indication that the larger peak could be considered multimodal with peaks occurring at ∼0, and slightly <1 (fig. 4). Interestingly, the lower peak (centered at approximately −2) in the SVMP distribution was composed exclusively of Agkistrodon contortrix and A. piscivorus lineages, suggesting that reliance on a particular subset of SVMP paralogs may be characteristic of the A. contortrix + A. piscivorus lineage. In SVSPs, the two apparent modes of the distribution appeared centered at approximately −0.5 and slightly <1 (fig. 4), though there was no apparent taxonomic pattern associated with either mode.
Fig. 4.
Density distributions of standardized MGD for the SVMP (a) and SVSP (b) gene families. Correlations between expression-weighted and unweighted standardized MGD are shown as insets with P-values and R2 values inferred by linear regression. Dashed red lines show the fitted slopes and solid black lines show the one-to-one line.
Under scenarios where SVMP and SVSP assemblages are evolutionarily constrained to emphasize either ancient orthologs or recent paralogs, we would expect distributions centered above or below zero, respectively. In contrast, the observed patterns suggest that the SVMP and SVSP evolution occurs through a combination of gene duplication, divergence, and loss rather than either extreme mechanisms of high duplication or high divergence (fig. 4). Moreover, the multimodal patterns of each distribution indicate that, whereas there is substantial variation in the diversity of assemblages, subsets of taxa exhibit especially similar or especially diverse SVMP and SVSP assemblages. Expression-weighted MGD was highly correlated with standardized MGD for both metrics (fig. 4), demonstrating that lineages did not emphasize the expression of more or less diverse paralogs in their total toxin assemblage.
Although we found no evidence of constraint on the genetic mechanisms for generating SD, it is possible that different mechanisms of generating diversity could facilitate broader diets. For example, more genetically diverse toxin assemblages might affect a wider phylogenetic diversity of prey, increasing DB. To test this possibility, we compared the MGD estimates (which represented more and less diverse toxin assemblages) to DB estimates for each lineage. However, we found no evidence for a relationship between DB and MGD (supplementary fig. S12, Supplementary Material online), indicating that the genetic diversity of toxin assemblages (i.e., emphasis on highly diverged vs. recently diverged paralogs) did not constrain the ecological function of venoms.
Discussion
Our results demonstrate that both SD and expression variation in toxin genes jointly shape variation in venom, a crucial adaptive trait related to DB in pitvipers. Previous studies have provided evidence for positive selection acting on toxin genes implicating the proteins they encode in trophic adaptions (Duda and Palumbi 1999; Li et al. 2005; Gibbs and Rossiter 2008; Sunagar and Moran 2015; Haney et al. 2016). Similarly, there is substantial indirect evidence for the role of expression variation in single toxins mediating trophic adaptations (Gibbs and Chiucchi 2011; Aird et al. 2015; Margres et al. 2016; Margres, Wray et al. 2017; Barua and Mikheyev 2019; Barua and Mikheyev 2020). Our study represents an advance by using comparative methods to simultaneously link the contribution of each molecular mechanism to phenotypic variation directly related to diet across diverse lineages. For certain key venom proteins, SD and expression appear to act in a hierarchical manner to generate the realized adaptive phenotype (whole venom composition). Diversity in protein sequence defines the fundamental functional sequence space for toxin proteins and expression variation brings about the realized toxin phenotype as a refined subset of sequence space. Such a model has been proposed to explain diversity in other venomous systems and variation in expression more broadly (Raser and O’Shea 2005; Lluisma et al. 2012). We suspect that a similar relationship will hold for other adaptive phenotypes whose function is driven by additive effects among component proteins.
The positive relationship between toxin SD and DB reinforces the idea that target-mediated interactions at the protein sequence level are a fundamental mechanism mediating predator–prey interactions through molecular phenotypes (Gibbs et al. 2020; Holding et al. 2021). Holding et al. (2021) demonstrated a correlation between overall toxin diversity and divergence in homologous venom targets involved in interactions with a single venom toxin (SVSPs). Our results build on this finding by demonstrating that both increased sequence and ED jointly underlie more diverse toxin compositions. A higher diversity of toxins may increase the number and type of physiological targets, and by extension, the number of physiologically distinct prey taxa that venom can affect (Davies and Arbuckle 2019). We suggest that these same mechanisms underlie positive correlations between venom and diet diversity that have been documented in other venomous animals such as snails and spiders (Phuong et al. 2016; Pekár et al. 2018).
We have modeled the relationship between DB, venom, and its genetic underpinning as a unidirectional genotype–phenotype relationship. This approach was effective for identifying how particular genetic mechanisms shape venom evolution but has limitations. In particular, path analyses cannot model bidirectional relationships as might be most appropriate in a feedback or coevolutionary system. This is potentially important because venoms that function primarily for prey capture likely evolve in complex, coevolutionary arms races with their prey in a variety of ecological scenarios (Barlow et al. 2009; Holding, Biardi, et al. 2016; Davies and Arbuckle 2019; Gibbs et al. 2020). Deciphering if and how prey characteristics like molecular resistance to venoms (Holding et al. 2018; Gibbs et al. 2020) shape snake venoms through coevolutionary interactions would be a valuable direction for future studies.
Our analysis of gene-family evolution in SVMP and SVSP paralogs shows no dominant mode of paralog duplication in achieving SD in toxin coding sequences. Instead, diverse toxin repertoires have emerged through the retention of deeply divergent paralogs, duplication, and comparatively minor divergence of paralogs, or a combination of these processes with equal likelihood. These findings are consistent with a previous study assessing expressed toxin assemblages in cone snails. Of the four species compared in cone snails (Chang and Duda 2014), two species expressed mostly similar paralogs (genetic underdispersion), one species expressed mostly divergent paralogs (genetic overdispersion), and one species fell between these extremes. Thus, in both snakes and cone snails, there is little constraint on the evolutionary pathway to achieving high SD in toxin genes—rather all pathways seem equally likely. Moreover, we found no association between the genetic diversity of toxin assemblages (MGD) and DB, indicating that having few, highly divergent paralogs versus many, less divergent paralogs did not have functional consequences for prey acquisition.
Given that venom targets basal physiological processes such as the coagulation cascade (Serrano 2013) and neurotransmission sites (Fry et al. 2009), it may be that relatively few amino acid substitutions can refine venom targeting for divergent prey tissues. The further divergence in more ancient paralogs may reflect the combined effects of neutral evolution (Aird et al. 2017) and refinements to protein function not tied to prey specificity, such as structural stability of the protein (Sunagar et al. 2014), neofunctionalization for novel physiological targets (Whittington et al. 2018), and modifications during pairwise coevolution to avoid inhibitor molecules of resistant prey (Holding, Biardi, et al. 2016; Margres, Bigelow, et al. 2017). Broadly, diet expansion appears possible through sequence variation derived from multiple possible pathways rather than any specific type of variation.
Importantly, the variation in modes of adaptions that we observed among different toxin families and the differences in their contribution to a complex phenotype demonstrate genomic heterogeneity in response to selective pressures associated with prey acquisition. In our study, the SVMP and SVSP toxins appear to influence DB through the maximization of toxin SD and ED. However, we did find some evidence of nonindependence of these mechanisms in SVSPs, where phylogenetic path analyses suggested direct interactions between SD and ED. Such a case may reflect scenarios, where differentially expressed toxins are experiencing differential rates of sequence evolution or cases where selection to increase expression leads to increased gene duplication and differentiation (Kondrashov and Kondrashov 2006; Kondrashov 2012; Aird et al. 2015; Margres, Bigelow, et al. 2017).
In contrast, the path analysis of PLA2s showed no support for a SD mediated relationship with diet. Rather, PLA2s showed a strong positive relationship between mean expression and DB, suggesting that an investment in PLA2 expression is associated with increased prey diversity. Why PLA2s exhibit this distinct relationship between diet and expression is not clear, but one possibility is that it reflects a broad functional efficacy of the same proteins across diverse taxa. PLA2s exhibit a wide range of functional effects including muscular and nervous system targeted neurotoxicity and myotoxicity (Gutiérrez and Lomonte 2013), which may be less specialized, but similarly effective among phylogenetically distinct prey groups. Thus, the role of PLA2s in shaping diet diversity might be better described by a mechanism whereby a given toxin or toxin family is broadly effective in a variety of scenarios at the cost of being less effective at targeting specific diet items. Alternatively, PLA2s may be especially effective against taxonomic groups that tend to be or are exclusively associated with broader diets, although evidence for this hypothesis is mixed and in need of further investigation (Lomonte et al. 2009).
The functions and effects of CTL diversity on diets remain unclear, as we found no evidence of an association between genetic variation and DB in this toxin family. The deviation of CTLs from other snake venom families is consistent with earlier tests comparing the relationship between DB and mRNA k-mer diversity among toxin families (Holding et al. 2021). Notably, CTLs are unique among snake venom toxins for functioning as multimeric heterodimers, which could impose unique restrictions on their evolvability or decouple a direct relationship between genetic and functional variation (Arlinghaus and Eble 2012; Eble 2019).
In conclusion, our study demonstrates the power of combining high-resolution transcriptomic datasets with comparative approaches to identify the molecular underpinnings of key adaptations in phylogenetically diverse nonmodel and emerging-model organisms. Our findings suggest both SD in protein-coding genes and how this diversity is regulated and ultimately expressed play key roles in mediating functional variation in the components of venom, but that the role of these mechanisms is not ubiquitous for all components. Molecular traits such as animal venoms, phytochemicals, and immune gene products are at the interface of antagonistic interactions among much of the planet’s biodiversity. Our study demonstrates that the genomic pathways to adaptive variation in these systems are as multifaceted and complex as the phenotypes themselves.
Materials and Methods
Bioinformatic Processing of Transcriptomes
We assembled and annotated venom-gland transcriptomes for 214 individuals from 68 rattlesnake and moccasin lineages used in Holding et al. (2021). All data processing was conducted using the Owens computing cluster at the Ohio Supercomputing Center (Center 1987). Briefly, raw sequence data were trimmed using TrimGalore! v.0.6.4 (Krueger 2015) and merged using PEAR v0.9.6 (Zhang et al. 2014). Merged reads were used to generate three transcriptome assemblies for each individual following the recommendations of (Holding et al. 2018). We used Trinity v.2.9.1 (Grabherr et al. 2011) and Seqman NGen 14 with default settings, and Extender v1.03 (Rokyta et al. 2012) with an overlap value of 120, a minimum seed quality of 30, replicates value of 20, and a minimum of 20 passes. These three assemblies were combined into a single master assembly and annotated with ToxCodAn (Nachtigall et al. 2021).
Annotated transcriptomes were subjected to several filters to reduce the inclusion of erroneously recovered transcripts. First, a custom python script, ChimeraKiller v.0.7.3 (https://github.com/masonaj157/ChimeraKiller) was used to filter out likely chimeric sequences based on the distribution of reads across each site in the coding region. Second, transcripts were filtered for incomplete coding regions and putatively premature stop codons. Third, we filtered out sequences with unreliable read coverage. These were defined as sequences with <10× coverage for >10% of the sequence. Finally, we removed transcripts from the four largest snake toxin families (CTLs, PLA2s, SVMPs, and SVSPs) with transcript per million (TPM) estimates <300, which may have been assembled due to barcode misassignment during sequencing. All python scripts used in transcriptome filtering steps are available on GitHub at https://github.com/masonaj157/Statistical_Analyses_For_Phylogenetic_Comparisons_of_North_American_Pitviper_Transcriptomes.
After filtering, transcripts were clustered at a 98% similarity using cd-hit-est v.4.8.1 (Fu et al. 2012) to cluster alleles or very recent paralogs (Hofmann et al. 2018; Strickland et al. 2018). This represented the final transcriptome assembly for each sample. To estimate transcript expression, merged reads for each individual were mapped to their final transcriptome using Bowtie2 (Langmead and Salzberg 2012) as implemented in RSEM v.1.3.3 (Li and Dewey 2011). At this stage, we excluded one sample, C. durissus SB0275, from downstream analysis because it had an unusually low number of raw reads which resulted in a low-quality transcriptome assembly.
Using the final transcriptome and estimated expression, we calculated three metrics characterizing genetic sources of complexity in venom toxins: (1) GC, (2) toxin amino acid SD, and (3) ED. We calculated GC of the transcriptome as the total number of unique toxin transcripts recovered in the final transcriptomes. We use GC as an estimate of the number of distinct loci present in a given sample. Because the venom phenotype’s interaction with prey is a function of protein composition, we characterized toxin SD through amino acid 20-mer content. For each individual, we translated toxins, counted all unique 20-mers (script available on the project GitHub), and summarized amino acid diversity with Shannon’s diversity index (H) converted to effective numbers of k-mers. We assume this measure captures the overall functional diversity in protein-coding sequences present in a transcriptome. Finally, to estimate ED, we calculated Shannon’s H per specimen treating toxins as “individuals” and TPM as “counts,” which were converted to effective numbers of transcripts. For this measure of ED, higher values represent more even expression across transcripts, and therefore, greater functional diversity. These metrics were then averaged among specimens belonging to the same lineage to attain lineage-level estimates that were used in subsequent analyses. Further details on the calculation of each index are provided in the supplementary Material, Supplementary Material online.
We assessed the possible influence of technical variations, such as variation in sequencing effort and transcriptome completeness, on toxin transcript recovery by testing for correlations between GC versus the number of reads and the total numbers of expressed transcripts with linear models implemented with the lm function in R. To further ensure that these technical sources of variation did not affect downstream analyses through phylogenetic biases, we also tested for an interaction between lineage and either the read numbers or total numbers of expressed transcripts on toxin GC with two linear models implemented in R and summarized with the “Anova” function of the car v.3.0-10 package (Fox and Weisberg 2019).
We tested whether our calculated variables for venom diversity exhibited evidence of phylogenetic signal as was found for the whole venom phenotype by testing for the significance of Blomberg’s K and lambda, two common metrics of phylogenetic signal. Blomberg’s K assesses the variance among species compared with the expected variance under Brownian motion, whereas lambda is a tree scaling parameter with an expected value of 0 if there is no correlation among species and 1 if correlation matches Brownian motion. For each variable, we assessed the phylogenetic signal and tested for a significant phylogenetic signal using the “phylosig” function of phytools (Revell 2012) specifying either “method = K” or “method = lambda” and “test = TRUE.”
Phylogenetic Path Analysis
To test for possible causal relationships between DB and molecular sources of venom variation, we evaluated a range of phylogenetic path models for the 30 pitvipers with reliable diet information (Holding et al. 2021) using the R package phylopath (van der Bijl 2018). We tested 10 models representing different hypotheses regarding the direct and indirect influences of GC, SD, ED (defined as above), and DB (as measured by the standardized MPD of prey—see Holding et al. 2021) (supplementary fig. S7, Supplementary Material online). We used MPD of prey as our measure of DB because Holding et al. (2021) found that this estimate of diet showed the strongest positive relationship to different measures of venom complexity likely because it incorporates information on functional diversity of venom targets in prey. Values for this index have a positive relationship with DB with higher values indicating broader diets. Generally, these models incorporate varying roles of SD and ED as directly or indirectly predicting DB, independently or in combination, whereas GC acted indirectly through these variables. This framework, where venom variables predict diet breath is consistent with a hierarchical “genotype → phenotype → ecological-outcome” framework (Barrett and Hoekstra 2011), which models how species adapt to their environments. Importantly, this model allows the cumulative variation of GC, SD, and ED cumulatively to predict DB, but should not be taken to imply directionality in the venom–diet association (supplementary methods, Supplementary Material online).
Because the cumulative sequence and expression diversity are partially a function of what genes are expressed, they covary with one another. To account for this covariance, we included the direct effects of GC on SD and ED in all tested models. A model which only included the effects of GC on SD and ED, but no relationship between the SD and ED on diet diversity was used as the null model to account for any consistent correlation that is otherwise unrelated to diet (supplementary fig. S7, Supplementary Material online). Likewise, because the effect of differential GC can only be realized in the venom phenotype through changes in toxin SD and/or expression, no models included a direct relationship between the GC and DB.
All path models were estimated under a lambda model of evolution and compared using CICc. The framework for CIC was proposed by Cardon et al. (2011) and has recently been established for use in the phylogenetic path analysis (von Hardenberg and Gonzalez-Voyer 2013; Voyer and Garamszegi 2014). Briefly, CICc is calculated using a model’s C statistic, a number of parameters, and a correction for small sample size (Voyer and Garamszegi 2014). Under this framework, models with the same numbers of variable relationships but different directionalities are expected to show similar statistical support, but their differing effect estimates may still be informative. Because a single model was not statistically preferred over all other models, we also estimated a weighted average model with weights determined from model likelihoods. All paths within comparably performing models (i.e., those within two CICc) were averaged. We also obtained confidence intervals for path coefficient estimates (partial regression coefficients standardized to the other independent variables) with 500 bootstraps. The parameters provided to the ‘phylo_path’ function were the predefined model set, the data frame of venom and DB variables, the calibrated phylogeny, and the model specification “model = lambda.” All other parameters were left as defaults.
In addition to performing the phylogenetic path analysis for the overall venom dataset (all toxin classes combined), we also examined variation among the patterns of evolution within four major toxin families: CTLs, PLA2s, SVMPs, and SVSPs which represent major components of venom in these snakes (Holding et al. 2021). For each family, we restricted the dataset to toxins assigned to that family based on ToxCodAn annotation and estimated GC, SD, and ED. Each family was subsequently tested with the phylogenetic path analysis using the same methods that had been applied to the whole dataset.
Variation in Expression
Phylogenetic path analyses found counterintuitive and conflicting results for the role of ED at the whole venom level compared with what was recovered for the SVMP and SVSP families. Because ED can be decomposed into the roles of richness (number of transcripts) and relative expression of each transcript, we hypothesized that differences in the number and expression of toxins in highly expressed toxin families would explain the trends observed in the path analyses. To assess how transcript numbers and expression varied among large, highly expressed toxin families, we compared transcript numbers and mean toxin expression in CTLs, PLA2s, SVMPs, and SVSPs. We then tested for a correlation between expression and DB in these families to identify the disproportionate drivers of ED.
First, to account for the compositional constraints of expression estimates, we performed a centered log-ratio (CLR) transformation of TPM data for each individual. The CLR transformed TPM values were then used in all subsequent comparisons of expression. We then calculated the mean expression of transcripts in the CTL, PLA2, SVMP, and SVSP families. For a few samples, no toxins were recovered for a particular gene family (i.e., CTLs, PLA2s, SVMPs, or SVSPs) and their toxin numbers and expression values were encoded as NA. As a failure to recover a toxin could occur because of stochastic variation in transcriptome assembly or our conservative approach to toxin filtering, such samples were excluded from the analysis of that gene family. To attain lineage-specific estimates, we averaged the number of expressed transcripts and mean expression of individuals in a phylogenetic lineage. We tested for the overall differences in the numbers of expressed toxins and mean toxin expression among toxin families with an ANOVA in R treating toxin family as the independent variable and lineage as a block variable. Differences among treatments were tested with Bonferroni corrected post hoc t-tests. Finally, to determine if any variation in expression was associated with DB, we tested for relationships between DB and mean toxin expression within each toxin family with a phylogenetic linear regression implemented with phylolm v.2.6 (Ho and Ane 2014).
Evolution of Genetic Diversity of SVMP and SVSP Paralogs
Our path analyses showed a direct relationship between toxin SD and DB. To explore how SD was generated at the gene level for these toxins, we used an approach proposed by Chang and Duda (2014), which uses community phylogenetics indices to characterize the diversity of a toxin assemblage against the total diversity of a gene family—in this case, the total diversity of SVMP or SVSP paralogs observed in Agkistrodon, Crotalus, and Sistrurus. As individual snakes normally express several SVMP and SVSP paralogs, metrics such as standardized MGD can be calculated for each gene family in each individual. These indices identify where on a continuum that ranges from a high divergence between distinct paralogs to a limited divergence between related paralogs, a given set of expressed transcripts falls. This permits an indirect but quantitative inference of the evolutionary processes in terms of gene family and sequence evolution.
To conduct these analyses on our data, we first compiled translated mRNA sequences for all recovered toxins in each family and generated a gene-family alignment using MUSCLE v3.8.1551 (Edgar 2004). We then generated separate maximum-likelihood gene-family phylogenies for the SVMP and SVSP gene families using iqtree (Nguyen et al. 2015). Evolutionary models were selected for each family using iqtree’s ModelFinder feature and we recovered branch support estimates with 1000 ultrafast bootstraps. These full gene-family phylogenies represented the full diversity of SVMPs and SVSPs observed among all Agkistrodon, Crotalus, and Sistrurus. Using these two trees, we calculated standardized MGD for the SVMP and SVSP gene families for each individual using the ses.mpd function in the ‘picante’ package in R (Kembel et al. 2010). The resultant standardized MGD values represented the relative diversity of SVMP or SVSP paralogs expressed by a given individual compared with the total diversity of SVMP or SVSP paralogs in Agkistrodon, Crotalus, and Sistrurus. To account for the possible role of expression variation in altering realized the diversity of toxin assemblages, we also calculated expression-weighted standardized MGD using the TPM values of each toxin as abundance estimates. Standardized and expression-weighted MGD values were then averaged across individuals for lineages with multiple representatives for lineage-level estimates of standardized MGD. Additional details on the calculation of MGD and weighted MGD are provided in the supplementary material, Supplementary Material online.
Using the standardized MGD values, we estimated whether expression weighting had a strong effect on altering diversity and we tested for a relationship between standardized MGD, SD, and DB. We tested for differences between the standardized and expression-weighted MGD with a standard linear regression and R2 estimate using the “lm” function in R. Because distributions appeared multimodal, we also tested whether each distribution was significantly different than 0 with a one-sided Wilcoxon signed rank test with the “wilcox.text” function in R. To determine if the genetic diversity of toxin assemblages was associated with venom evolution, we then tested for relationships between standardized MGD and SD with phylogenetic linear regression using the ‘phylolm’ package in R.
Supplementary Material
Acknowledgments
This study was funded by the National Science Foundation (DEB 1638872 to H.L.G., DEB 1638879 and DEB 1822417 to C.L.P., and DEB 1638902 to D.R.R.). We thank Matthew Hahn and Samarth Mathur for their comments on the manuscript. We gratefully acknowledge the Ohio Supercomputing Center which provided the high-performance computing resources used in this study. Animal icons used in figures were retreived from PhyloPic and were originally provided by Bill Bouton, T. Michael Keesey, Steven Traver, Beth Reinke, Natasha Vitek, and Blair Perry. Bill Bouton and T. Michael Keesey graciously granted permission to use the snake icon presented in fig. 1.
Data Availability
The data underlying this article are available in the article or on the GenBank SRR and SRA databases under the accession numbers provided in supplemental tables S1 and S2, Supplementary Material online. The data on the metrics of phylogenetic diet complexity were collected from and are available in Holding et al. (2021). Copies of the input data files and R script used for data analysis are available on GitHub at: https://github.com/masonaj157/Statistical_Analyses_For_Phylogenetic_Comparisons_of_North_American_Pitviper_Transcriptomes.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
References
- Aird SD, Aggarwal S, Villar-Briones A, Tin MM-Y, Terada K, Mikheyev AS. 2015. Snake venoms are integrated systems, but abundant venom proteins evolve more rapidly. BMC Genom. 16:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird SD, Arora J, Barua A, Qiu L, Terada K, Mikheyev AS. 2017. Population genomic analysis of a pitviper reveals microevolutionary forces underlying venom chemistry. Genome Biol Evol. 9:2640–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuckle K. 2020. From molecules to macroevolution: venom as a model system for evolutionary biology across levels of life. Toxicon X 6:100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlinghaus FT, Eble JA. 2012. C-type lectin-like proteins from snake venoms. Toxicon 60:512–519. [DOI] [PubMed] [Google Scholar]
- Barlow A, Pook CE, Harrison RA, Wüster W. 2009. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc R Soc B 276:2443–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Hoekstra HE. 2011. Molecular spandrels: tests of adaptation at the genetic level. Nat Rev Genet. 12:767–780. [DOI] [PubMed] [Google Scholar]
- Barua A, Mikheyev AS. 2019. Many options, few solutions: over 60 million years snakes converged on a few optimal venom formulations. Mol Biol Evol. 36:1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua A, Mikheyev AS. 2020. Toxin expression in snake venom evolves rapidly with constant shifts in evolutionary rates. Proc R Soc B Biol Sci. 287:20200613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F, Picao-Osorio J, Dubois C, Félix MA. 2020. A broad mutational target explains a fast rate of phenotypic evolution. Elife 9:1–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon M, Loot G, Grenouillet G, Blanchet S. 2011. Host characteristics and environmental factors differentially drive the burden and pathogenicity of an ectoparasite: a multilevel causal analysis. J Anim Ecol. 80:657–667. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2005. Evolution at two levels: on genes and form. PLoS Biol. 3:e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Huttley GA, Wüster W. 2012. Dynamic evolution of venom proteins in squamate reptiles. Nat Commun. 3:1–10. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Petras D, Card DC, Suranse V, Mychajliw AM, Richards D, Koludarov I, Albulescu L-O, Slagboom J, Hempel B-F, et al. . 2019. Solenodon genome reveals convergent evolution of venom in eulipotyphlan mammals. Proc Natl Acad Sci U S A. 116:25745–25755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Wagstaff SC, Wüster W, Cook DAN, Bolton FMS, King SI, Pla D, Sanz L, Calvete JJ, Harrison RA. 2014. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc Natl Acad Sci U S A. 111:9205–9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. 2013. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 28:219–229. [DOI] [PubMed] [Google Scholar]
- Chak STC, Harris SE, Hultgren KM, Jeffery NW, Rubenstein DR. 2021. Eusociality in snapping shrimps is associated with larger genomes and an accumulation of transposable elements. Proc Natl Acad Sci U S A. 118:e2025051118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Duda TF. 2012. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol Biol Evol. 29:2019–2029. [DOI] [PubMed] [Google Scholar]
- Chang D, Duda TF. 2014. Application of community phylogenetic approaches to understand gene expression: differential exploration of venom gene space in predatory marine gastropods. BMC Evol Biol. 14:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E-L, Arbuckle K. 2019. Coevolution of snake venom toxic activities and diet: evidence that ecological generalism favours toxicological diversity. Toxins 11:711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Palumbi SR. 1999. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc Natl Acad Sci U S A. 96:6820–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Palumbi SR. 2004. Gene expression and feeding ecology: evolution of piscivory in the venomous gastropod genus Conus. Proc R Soc Lond Ser B Biol Sci. 271:1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Remigio EA. 2008. Variation and evolution of toxin gene expression patterns of six closely related venomous marine snails. Mol Ecol. 17:3018–3032. [DOI] [PubMed] [Google Scholar]
- Eble JA. 2019. Structurally robust and functionally highly versatile—C-type lectin (-related) proteins in snake venoms. Toxins 11:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, Weisberg S. 2019. An {R} companion to applied regression. 3rd ed. Thousand Oaks (CA): Sage. Available from: https://socialsciences.mcmaster.ca/jfox/Books/Companion/. [Google Scholar]
- Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JDA, King GF, Nevalainen TJ, Norman JA, Lewis RJ, Norton RS, et al. . 2009. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 10:483–511. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau KL, Haney RA, Schwager EE, Wierschin T, Stanke M, Richards S, Garb JE. 2017. House spider genome uncovers evolutionary shifts in the diversity and expression of black widow venom proteins associated with extreme toxicity. BMC Genom. 18:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs HL, Chiucchi JE. 2011. Deconstructing a complex molecular phenotype: population-level variation in individual venom proteins in eastern massasauga rattlesnakes (Sistrurus c. catenatus). J Mol Evol. 72:383–397. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Rossiter W. 2008. Rapid evolution by positive selection and gene gain and loss: PLA2 venom genes in closely related Sistrurus rattlesnakes with divergent diets. J Mol Evol. 66:151–166. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Sanz L, Pérez A, Ochoa A, Hassinger ATB, Holding ML, Calvete JJ. 2020. The molecular basis of venom resistance in a rattlesnake-squirrel predator-prey system. Mol Ecol. 29:2871–2888. [DOI] [PubMed] [Google Scholar]
- Giorgianni MW, Dowell NL, Griffin S, Kassner VA, Selegue JE, Carroll SB. 2020. The origin and diversification of a novel protein family in venomous snakes. Proc Natl Acad Sci U S A. 117:10911–10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. . 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignard MS, Crawley MJ, Kovalenko D, Nichols RA, Trimmer M, Leitch AR, Leitch IJ. 2019. Interactions between plant genome size, nutrients and herbivory by rabbits, molluscs and insects on a temperate grassland. Proc Biol Sci. 286:20182619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez JM, Lomonte B. 2013. Phospholipases A2: unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon 62:27–39. [DOI] [PubMed] [Google Scholar]
- Haney RA, Clarke TH, Gadgil R, Fitzpatrick R, Hayashi CY, Ayoub NA, Garb JE. 2016. Effects of gene duplication, positive selection, and shifts in gene expression on the evolution of the venom gland transcriptome in widow spiders. Genome Biol Evol. 8:228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LST, Ane C. 2014. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst Biol. 63:397–408. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61:995–1016. [DOI] [PubMed] [Google Scholar]
- Hofmann EP, Rautsaw RM, Strickland JL, Holding ML, Hogan MP, Mason AJ, Rokyta DR, Parkinson CL. 2018. Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Sci Rep. 8:15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding ML, Biardi JE, Gibbs HL. 2016. Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc R Soc B Biol Sci. 283:20152841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding ML, Drabeck DH, Jansa SA, Gibbs HL. 2016. Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integr Comp Biol. 56:1032–1043. [DOI] [PubMed] [Google Scholar]
- Holding M, Margres M, Mason A, Parkinson C, Rokyta D. 2018. Evaluating the performance of de novo assembly methods for venom-gland transcriptomics. Toxins 10:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding ML, Strickland JL, Rautsaw RM, Hofmann EP, Mason AJ, Hogan MP, Nystrom GS, Ellsworth SA, Colston TJ, Borja M, et al. . 2021. Phylogenetically diverse diets favor more complex venoms in North American pitvipers. Proc Natl Acad Sci U S A. 118:2015579118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Sackton TB, Edwards SV, Liu JS. 2019. Bayesian detection of convergent rate changes of conserved noncoding elements on phylogenetic trees. Mol Biol Evol. 36:1086–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc R Soc B Biol Sci. 279:5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Kondrashov AS. 2006. Role of selection in fixation of gene duplications. J Theor Biol. 239:141–151. [DOI] [PubMed] [Google Scholar]
- Krueger F. 2015. Trim Galore! : A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Available from: https://github.com/FelixKrueger/TrimGalore
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barghi N, Lu A, Fedosov AE, Bandyopadhyay PK, Lluisma AO, Concepcion GP, Yandell M, Olivera BM, Safavi-Hemami H. 2017. Divergence of the venom exogene repertoire in two sister species of Turriconus. Genome Biol Evol. 9:2211–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fry BG, Kini RM. 2005. Putting the brakes on snake venom evolution: the unique molecular evolutionary patterns of Aipysurus eydouxii (Marbled Sea Snake) phospholipase A 2 toxins. Mol Biol Evol. 22:934–941. [DOI] [PubMed] [Google Scholar]
- Lluisma AO, Milash BA, Moore B, Olivera BM, Bandyopadhyay PK. 2012. Novel venom peptides from the cone snail Conus pulicarius discovered through next-generation sequencing of its venom duct transcriptome. Mar Genomics 5:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte B, Angulo Y, Sasa M, María Gutiérrez J. 2009. The phospholipase A 2 homologues of snake venoms: biological activities and their possible adaptive roles. Protein Pept Lett. 16:860–876. [DOI] [PubMed] [Google Scholar]
- Margres MJ, Bigelow AT, Lemmon EM, Lemmon AR, Rokyta DR. 2017. Selection to increase expression, not sequence diversity, precedes gene family origin and expansion in rattlesnake venom. Genetics 206:1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres MJ, Rautsaw RM, Strickland JL, Mason AJ, Schramer TD, Hofmann EP, Stiers E, Ellsworth SA, Nystrom GS, Hogan MP, et al. . 2021. The Tiger Rattlesnake genome reveals a complex genotype underlying a simple venom phenotype. Proc Natl Acad Sci U S A. 118:e2014634118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres MJ, Wray KP, Hassinger ATB, Ward MJ, McGivern JJ, Moriarty Lemmon E, Lemmon AR, Rokyta DR. 2017. Quantity, not quality: rapid adaptation in a polygenic trait proceeded exclusively through expression differentiation. Mol Biol Evol. 34:3099–3110. [DOI] [PubMed] [Google Scholar]
- Margres MJ, Wray KP, Seavy M, McGivern JJ, Herrera ND, Rokyta DR. 2016. Expression differentiation is constrained to low-expression proteins over ecological timescales. Genetics 202:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigorta UM, Rodríguez JA, Gibson G, Navarro A. 2018. Replicability and prediction: lessons and challenges from GWAS. Trends Genet. 34:504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtigall PG, Rautsaw RM, Ellsworth SA, Mason AJ, Rokyta DR, Parkinson CL, Junqueira-de-Azevedo ILM. 2021. ToxCodAn: a new toxin annotator and guide to venom gland transcriptomics. Brief Bioinform. 22:bbab095. [DOI] [PubMed] [Google Scholar]
- Nagy LG, Merényi Z, Hegedüs B, Bálint B. 2020. Novel phylogenetic methods are needed for understanding gene function in the era of mega-scale genome sequencing. Nucleic Acids Res. 48:2209–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohio Supercomputing Center . 1987. Ohio Supercomputer Center. Available from: http://osc.edu/ark:/19495/f5s1ph73.
- Ohno S. 1970. Evolution by gene duplication: Springer Science & Business Media. [Google Scholar]
- Pease JB, Haak DC, Hahn MW, Moyle LC. 2016. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14:e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekár S, Bočánek O, Michálek O, Petráková L, Haddad CR, Šedo O, Zdráhal Z. 2018. Venom gland size and venom complexity—essential trophic adaptations of venomous predators: A case study using spiders. Mol. Ecol. 27:4257–4269. [DOI] [PubMed] [Google Scholar]
- Phuong MA, Mahardika GN, Alfaro ME. 2016. Dietary breadth is positively correlated with venom complexity in cone snails. BMC Genom. 17:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. 2005. Noise in gene expression: origins, consequences, and control. Science 309:2010–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausher MD, Delph LF. 2015. Commentary: When does understanding phenotypic evolution require identification of the underlying genes? Evolution 69:1655–1664. [DOI] [PubMed] [Google Scholar]
- Rautsaw RM, Hofmann EP, Margres MJ, Holding ML, Strickland JL, Mason AJ, Rokyta DR, Parkinson CL. 2019. Intraspecific sequence and gene expression variation contribute little to venom diversity in sidewinder rattlesnakes (Crotalus cerastes). Proc R Soc B 286:20190810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigio EA, Duda TF. 2008. Evolution of ecological specialization and venom of a predatory marine gastropod. Mol Ecol. 17:1156–1162. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3:217–223. [Google Scholar]
- Rockman MV. 2012. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta DR, Lemmon AR, Margres MJ, Aronow K. 2012. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genom. 13:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta DR, Margres MJ, Calvin K. 2015. Post-transcriptional mechanisms contribute little to phenotypic variation in snake venoms. G3 (Bethesda) 5:2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta DR, Wray KP, McGivern JJ, Margres MJ. 2015. The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the timber rattlesnake (Crotalus horridus). Toxicon 98:34–48. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Grayson P, Cloutier A, Hu Z, Liu JS, Wheeler NE, Gardner PP, Clarke JA, Baker AJ, Clamp M, et al. . 2019. Convergent regulatory evolution and loss of flight in paleognathous birds. Science 364:74–78. [DOI] [PubMed] [Google Scholar]
- Sanggaard KW, Bechsgaard JS, Fang X, Duan J, Dyrlund TF, Gupta V, Jiang X, Cheng L, Fan D, Feng Y, et al. . 2014. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun. 5:3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schield DR, Card DC, Hales NR, Perry BW, Pasquesi GM, Blackmon H, Adams RH, Corbin AB, Smith CF, Ramesh B, et al. . 2019. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 29:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano SMT. 2013. The long road of research on snake venom serine proteinases. Toxicon 62:19–26. [DOI] [PubMed] [Google Scholar]
- Smith SD, Pennell MW, Dunn CW, Edwards SV. 2020. Phylogenetics is the new genetics (for most of biodiversity). Trends Ecol Evol. 35:415–425. [DOI] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution? Evolution 62:2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland J, Mason A, Rokyta D, Parkinson C, Strickland JL, Mason AJ, Rokyta DR, Parkinson CL. 2018. Phenotypic variation in Mojave Rattlesnake (Crotalus scutulatus) venom is driven by four toxin families. Toxins 10:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K, Casewell NR, Varma S, Kolla R, Antunes A, Moran Y. 2014. Deadly innovations: unraveling the molecular evolution of animal venoms. In: Gopalakrishnakone P, Calvete JJ, editors. Venom genomics and proteomics. Springer. p. 1–23. [Google Scholar]
- Sunagar K, Moran Y. 2015. The rise and fall of an evolutionary innovation: contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 11:e1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K, Morgenstern D, Reitzel AM, Moran Y. 2016. Ecological venomics: how genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J Proteomics. 135:62–72. [DOI] [PubMed] [Google Scholar]
- Tanksley SD. 1993. Mapping polygenes. Annu Rev Genet. 27:205–233. [DOI] [PubMed] [Google Scholar]
- van der Bijl W. 2018. Phylopath: easy phylogenetic path analysis in R. PeerJ 2018:e4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hardenberg A, Gonzalez-Voyer A. 2013. Disentangling evolutionary cause-effect relationships with phylogenetic confirmatory path analysis. Evolution 67:378–387. [DOI] [PubMed] [Google Scholar]
- Voyer AG, Garamszegi LZ. 2014. An introduction to phylogenetic path analysis. Modern phylogenetic comparative methods and their application in evolutionary biology. Springer Berlin Heidelberg. p. 201–229. [Google Scholar]
- Whittington AC, Mason AJ, Rokyta DR. 2018. A single mutation unlocks cascading exaptations in the origin of a potent pitviper neurotoxin. Mol Biol Evol. 35:887–898. [DOI] [PubMed] [Google Scholar]
- Wong ESW, Belov K. 2012. Venom evolution through gene duplications. Gene 496:1–7. [DOI] [PubMed] [Google Scholar]
- Zancolli G, Calvete JJ, Cardwell MD, Greene HW, Hayes WK, Hegarty MJ, Herrmann HW, Holycross AT, Lannutti DI, Mulley JF, et al. . 2019. When one phenotype is not enough: divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc R Soc B Biol Sci. 286:20182735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancolli G, Casewell NR. 2020. Venom systems as models for studying the origin and regulation of evolutionary novelties. Mol Biol Evol. 37:2777–2790. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article or on the GenBank SRR and SRA databases under the accession numbers provided in supplemental tables S1 and S2, Supplementary Material online. The data on the metrics of phylogenetic diet complexity were collected from and are available in Holding et al. (2021). Copies of the input data files and R script used for data analysis are available on GitHub at: https://github.com/masonaj157/Statistical_Analyses_For_Phylogenetic_Comparisons_of_North_American_Pitviper_Transcriptomes.