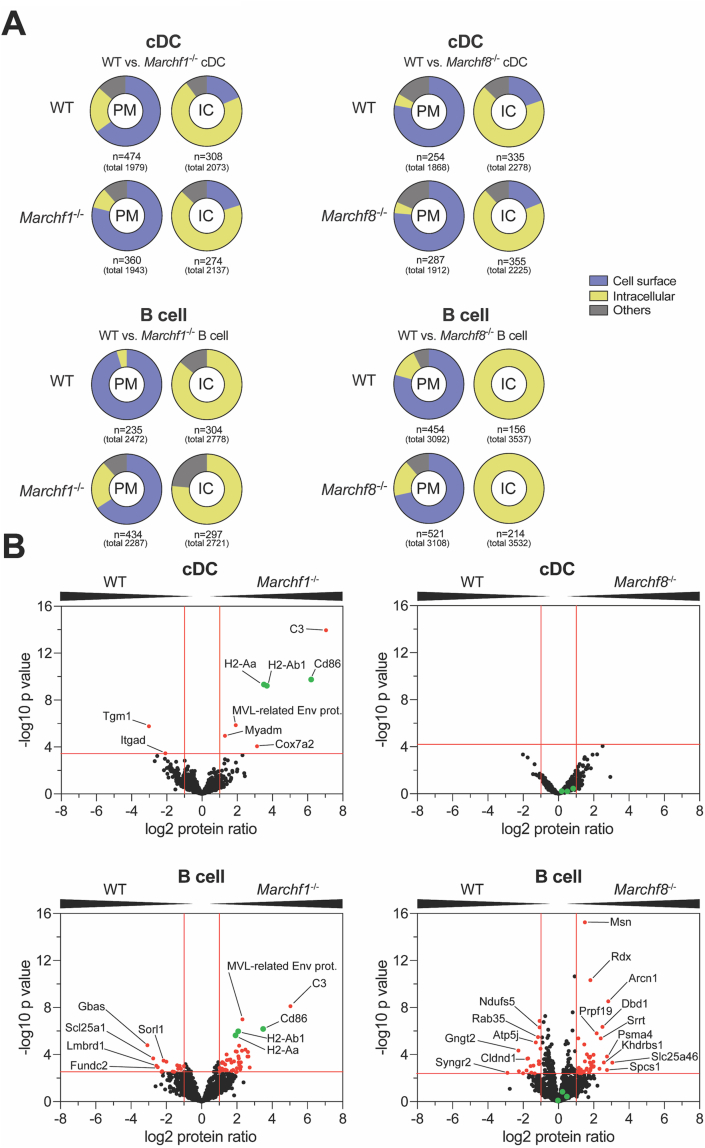
Fig. 5.
Proteomic analysis of differentially expressed proteins in the plasma membrane between WT and Marchf1-/-or Marchf8-/-cDCs and B cells. PM fractions were purified from PNS of mAb surface stained cDCs and B cells via magnetic immunoaffinity and analyzed by semi-quantitative mass spectrometry from three biological replicates (in total 3x 8 samples; WT vs. Marchf1-/- and WT vs. Marchf8-/- cDCs as well as WT vs. Marchf1-/- and WT vs. Marchf8-/- B cells). The remaining compartments (mitochondria, endosomes, etc.) sedimented from the PNS of homogenized cells following PM fraction retrieval was termed intracellular compartment (IC). (A) Enrichment analysis (performed using the function enricher included in the Bioconductor clusterProfiler package (Yu et al., 2012) of detected proteins via MS from PM or IC fractions from cDCs and B cells of WT versus Marchf1-/- and WT versus Marchf8-/- mice. Only experimentally verified Gene Ontology (GO) annotations were used (n = XY) from all detected proteins (total number in parenthesis), and grouped into categories of ‘Cell surface’, ‘Intracellular Compartment (IC)’ and ‘others’. ‘Cell surface’ category included the GO terms ‘plasma membrane’, ‘external side of plasma membrane’ and ‘cell surface’ among others, while the categories ‘Intracellular Compartment (IC)’ and ‘others’ included GO terms such as ‘mitochondrial membrane’ and ‘endoplasmic reticulum’ as well as ‘myelin sheath’, respectively. A detailed list of all annotated GO terms of all fractions can be found in Supplementary Fig. 4. (B) Detection of differentially expressed proteins in the PM fraction of cDCs and B cells of WT versus Marchf1-/- and WT versus Marchf8-/- mice. Equivalent amounts of PM fractions (based on cell count) of three biological replicates were analyzed by mass spectrometry and semi-quantitative proteomics in three technical replicates, for which the foldchange of the label-free quantification (LFQ) peptide peak intensities of each protein was used. Proteins detected in both WT and Marchf1-/- or Marchf8-/- cDCs or B cells were displayed in volcano plots (1020 proteins for WT vs. Marchf1-/- cDCs, 922 proteins for WT vs. Marchf8-/- cDCs, 1275 proteins for WT vs. Marchf1-/- B cells and 1819 proteins for WT vs. Marchf8-/- B cells) with differentially expressed proteins [red dots] identified based on two-fold ratio (log2 protein ratio >1 or <1) and significance (5% FDR) across three biological replicates, each measured in technical triplicates. The known MARCH1 substrates, MHC II (H2-Aa and H2-Ab1) and CD86 in B cells and cDCs, are highlighted in green in each volcano plot.