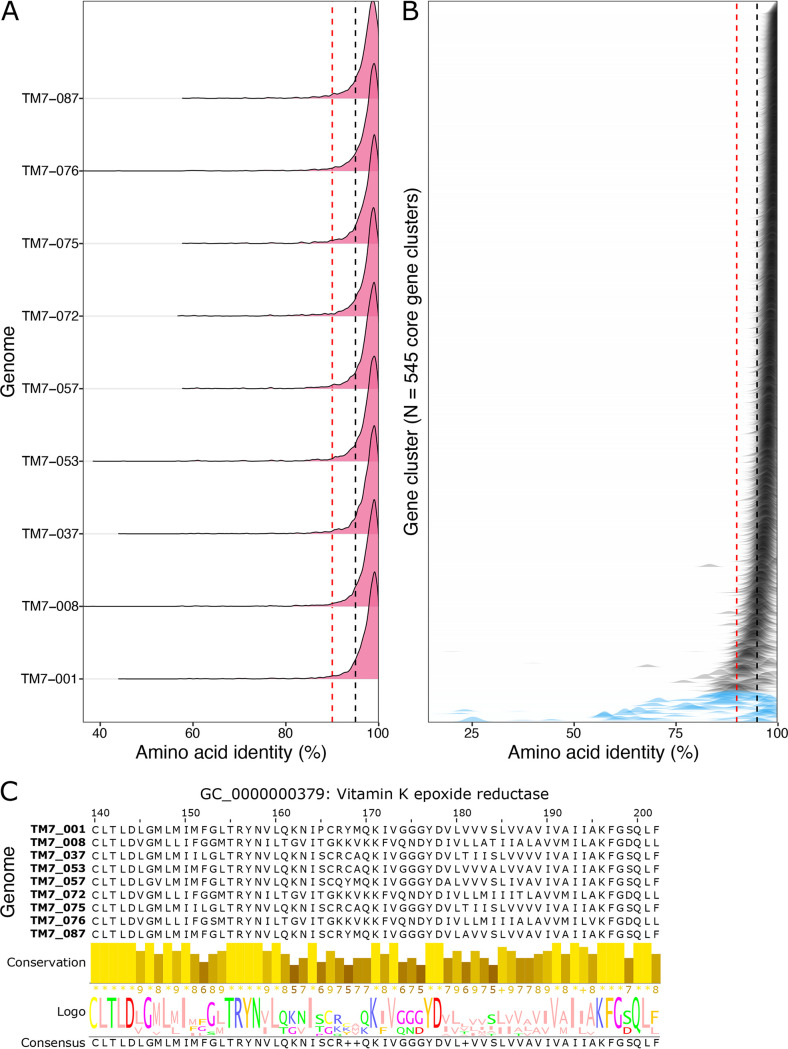
FIG 6.
Pairwise amino acid identity for HMT-352 core genes. Sequence identity was computed by gene cluster for all pairwise combinations of gene sequences for all nine genomes. Black and red dotted lines mark the 95% and 90% thresholds. (A) Density plots of pairwise amino acid sequence identity (x axis) by genome. Each density plot summarizes the observed amino acid sequence similarities compared to the other 8 genomes. (B) Density of observed percent identities by gene cluster, ordered by mean percent identity. Each distribution represents a different gene cluster. (C) Alignment and logo plot of a representative window of the vitamin K epoxide reductase gene. The height and shading intensity of the bars below the alignment show the conservation of each residue across the different genomes, while the logo plot below represents the frequency of the observed residues by the letter size.