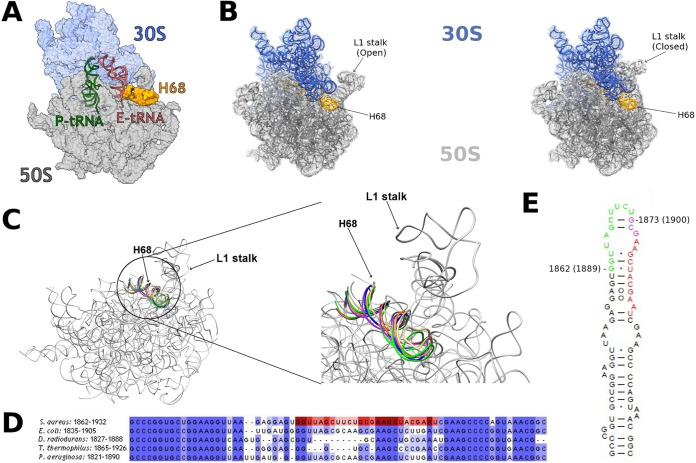
FIG 1.
Location and orientation of rRNA H68 in bacterial ribosomes. (A) The structure of S. aureus ribosome complex (SA70S) highlighting the locations of the rRNA H68 (orange) at the surface of the 50S (gray) subunit. The 30S subunit, P-tRNA, and E-tRNA are shown in light blue, green, and red, respectively. (B) The L1 stalk in opened (PDB ID 4V9H) and closed (PDB ID 4V9D) conformations. The 50S and the 30S are represented in gray and blue, respectively. (C) Overlay of H68 in various structures of bacterial ribosome. S. aureus 50S (SA50S) is shown in gray (PDB ID 6HMA); E. coli (PDB ID 4V9H), T. thermophilus (PDB ID 6CFK), D. radiodurans (PDB ID 4WFN), and P. aeruginosa (PDB ID 6SPF) are shown in tan, purple, orange, blue, and green, respectively. (D) Sequence alignment of H68 in various bacterial organisms. The regions targeted with antisense oligomers are marked in red. The image was created using Jalview (47). (E) Two-dimensional structure of S. aureus H68. The regions targeted with antisense oligomers are marked in green (1862 series) and red (1873 series) with the number of the first residue of each series according to E. coli numbering (S. aureus numbering is shown in parenthesis). The nucleotides targeted by both series are marked in purple.