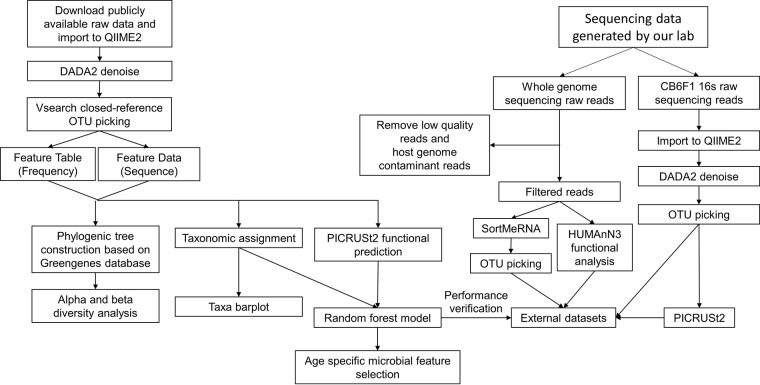
FIG 1.
Bioinformatic analysis pipeline. The raw 16S rRNA gene sequencing data were downloaded and imported into QIIME2 for data processing. DADA2 was used to denoise reads. OTU picking was performed by the Vsearch closed-reference method. The resulting feature table and feature data were then used for phylogenic tree construction and taxonomy annotation, followed by diversity analysis and taxon bar plot visualization. Functional prediction was performed by PICRUSt2. A random forest classifier was established based on predicted functional pathways to differentiate young- and old-age gut microbiomes and identify aging-specific metabolic features. The random forest classifier was then verified by two external data sets generated by us. The CB6F1 16S rRNA gene sequencing data were analyzed according to the same analysis pipeline. The whole-genome sequencing data were first filtered to remove low-quality and host genome contaminant reads, followed by SortMeRNA and OTU picking for taxonomy characterization and HUMAnN3 for functional analysis.