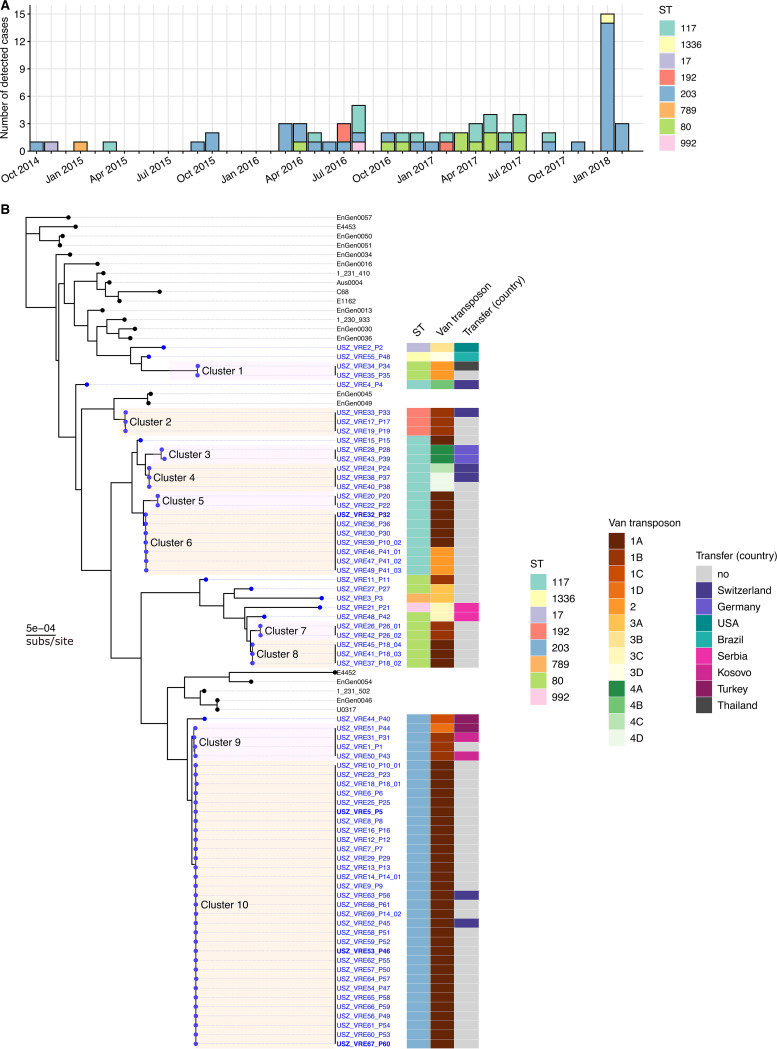
FIG 1.
Epidemiology and population structure of VREfm at the University Hospital of Zurich (USZ). (A) All VREfm cases detected at the USZ between October 2014 and March 2018. The colors represent the multilocus sequence type (ST) of each isolate. (B) Maximum-likelihood phylogenetic tree based on the core gene alignment (1,546,174 bp), rooted using the clade regrouping EnGen0051, EnGen0050, and EnGen0057, with E4453 as an outgroup. The tips of the phylogeny marked with black dots correspond to 21 isolates collected worldwide (2) (Data Set S1, Tab 2). Those marked with blue dots correspond to the 69 isolates of this study. The labels of the latter indicate the isolation location, USZ, the unique isolate identifier (ID), VRExx, and the ID of the patient from whom it was isolated, Pxx. In the few cases in which multiple isolates were derived from the same patient, the last number is the index of each isolate within the longitudinal series. The four isolates with boldface labels were additionally subjected to long-read sequencing to complete their assembly. Clusters, shaded with alternating colors for a clear visualization, were defined based on distances in this core gene alignment (Data Set S1, Tabs 3 and 4). The first column of the color map shows the ST of each isolate. The second column shows the variant of the vancomycin transposon. Three different types of transposon Tn1546, carrying the vanA genes, are labeled 1 to 3, and the subtypes they comprise further distinguished with letters and each highlighted with a different shade ranging from dark brown to light yellow. The transposon Tn1549 carrying the vanB genes is labeled 4 and the four subtypes it contains further distinguished with letters and highlighted with green shades. The third column of the color map shows which patients were admitted at USZ upon transfer from another hospital and the corresponding country of the hospital.