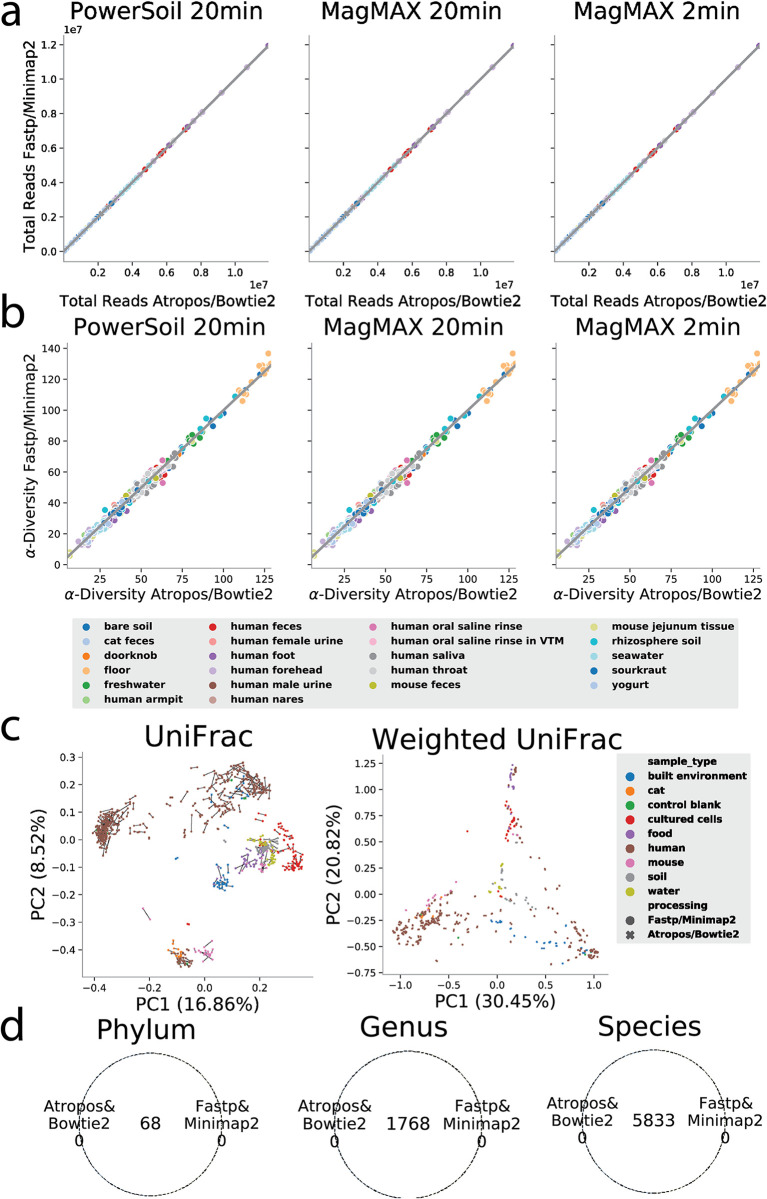
FIG 2.
When comparing broad sets of extraction kits and sample types, Minimap2/Fastp processing results do not differ in biological interpretation compared to current processing methods. (a and b) Comparison of total reads passing the filter (a) and Faith's phylogenetic diversity (b) for Fastp/Minimap2 (y axes) and Atropos/Bowtie2 (x axes) colored by sample type. (c) Principal coordinate analysis (PCoA) on unweighted (left) and weighted (right) UniFrac compared between Fastp/Minimap2 (circles) and Atropos/Bowtie2 (cross) colored by sample source environment. (d) Comparison of shared features between processing methods fastp/Minimap2 and Atropos/Bowtie2 at the phylum, genus, and species taxonomic levels.