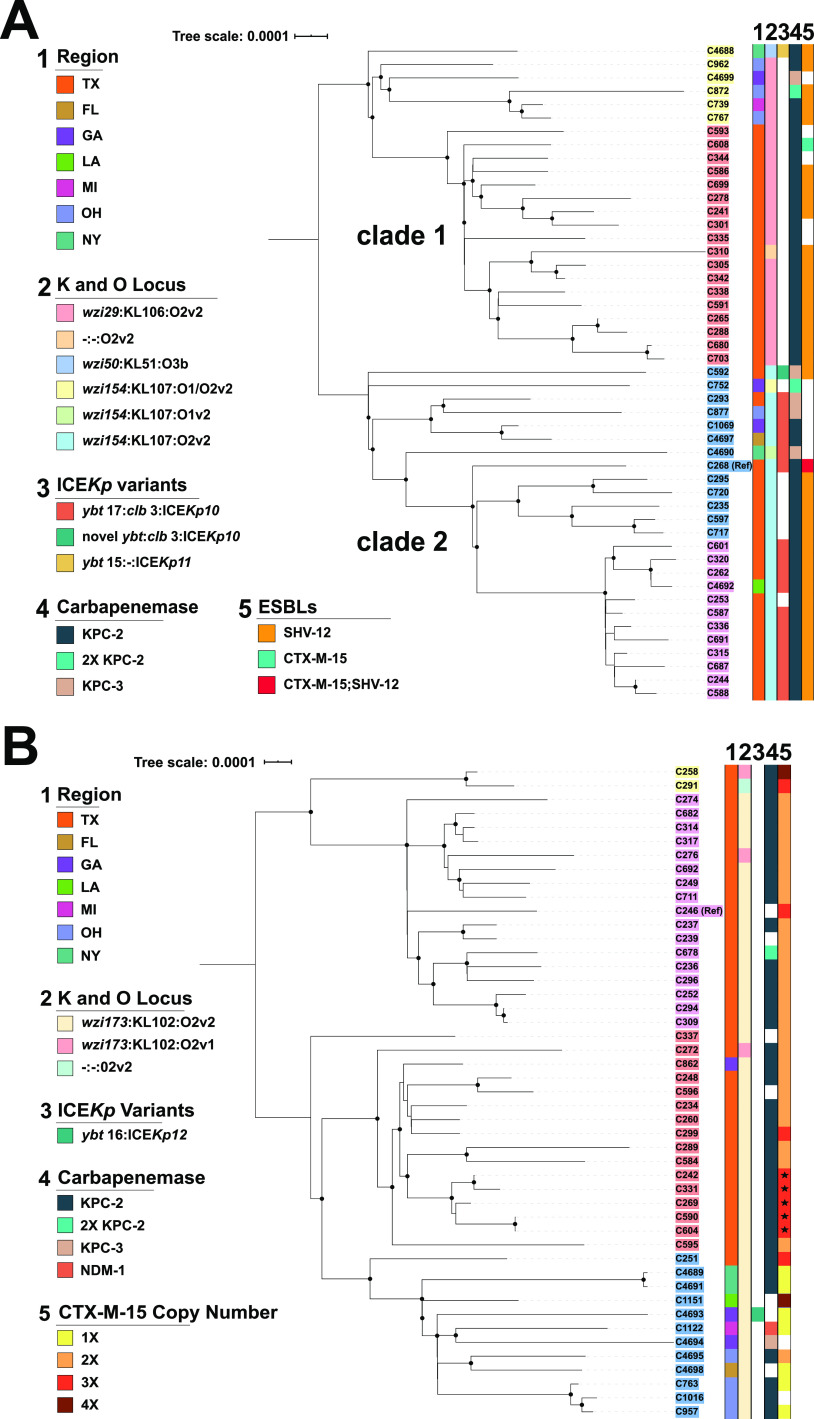
FIG 2.
Core SNP-inferred Maximum Likelihood (ML) phylogenomic trees of CG258 and CG307 isolates using a midpoint rooted tree. Internal node bootstrap values of ≥95% are denoted as black circles. Isolate label backgrounds indicate hierarchical clustering groups identified using Bayesian analysis of population structure for each respective sublineage. Legend designations are as follows: (1) region from which isolate was collected, (2) capsular synthesis/lipopolysaccharide (LPS) allele type, (3) ICEKp variant with yersiniabactin and colibactin gene cluster lineages listed, (4) carbapenemase carriage status, (5) ESBL carriage (CG258)/blaCTX-M-15 copy number (CG307). (A) CG258 core SNP phylogeny using a reference-based alignment with a C268 isolate. (B) CG307 core SNP phylogeny using reference-based alignment with a C246 isolate. Stars in the blaCTX-M-15 copy number column (i.e., column 5) indicate an isolate with one truncated blaCTX-M-15 copy.