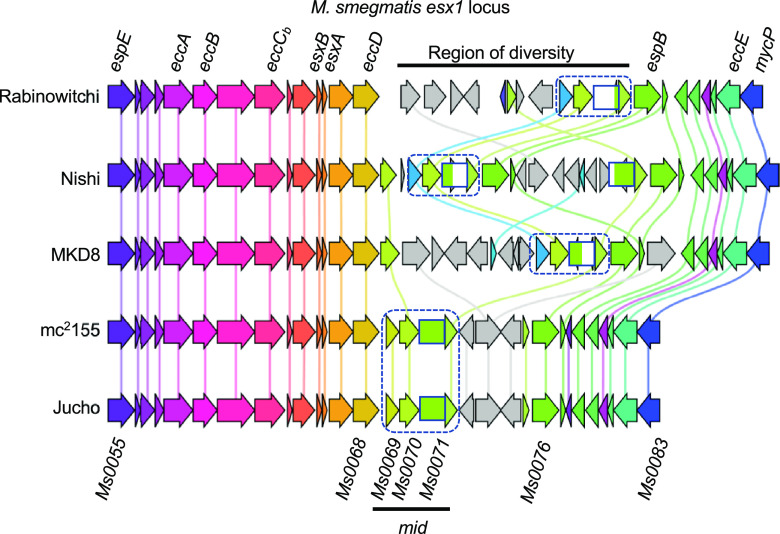
FIG 5.
Comparative gene analysis of the esx1 regions of the five M. smegmatis genomes. The schematic shows the overall collinearity and conservation of esx1 while highlighting the diversity of the mid region. The non-mid esx1 genes (Msmeg0055–0068 and Msmeg0076–0083) encode highly conserved proteins (>96.6% amino acid identity). However, proteins encoded by genes between these regions are poorly conserved (<30% amino acid identity) and include gene rearrangements, duplications, and multiple insertion sequence (IS) elements (mid genes within this region are boxed [Msmeg0069–0071]). Remarkably, mc2155 and Jucho are identical throughout the region, with 2 nucleotide differences from mc2155 in Msmeg0067 (resulting in an Arg-to-Pro amino acid change) and eccE (a silent C-to-T nucleotide substitution). Comparisons of esx1-encoded proteins were generated by Clinker using a best-BLAST-hit approach to identify orthologous genes at the 5′ and 3′ ends of each region (44). Orthologous genes are color-coded, and ISs and remnants of ISs are shaded in gray. Vertical lines drawn in the same color connect homologs. Note that the low amino acid conservation in the N terminus of Msmeg0071 prevented Clinker from identifying the complete gene, which we indicate here with a green striped box. Similarly, the low conservation of Msmeg0069 resulted in two classifications, identical in mc2155 and Jucho (olive arrow) and related but depicted in blue in Rabinowitchi, Nishi, and MKD8, immediately upstream of Msmeg0070–0071 orthologs. The names of conserved esx genes are indicated at the top of the alignment, and M. smegmatis numerical gene identifiers are shown at the bottom for reference.