Fig. 3.
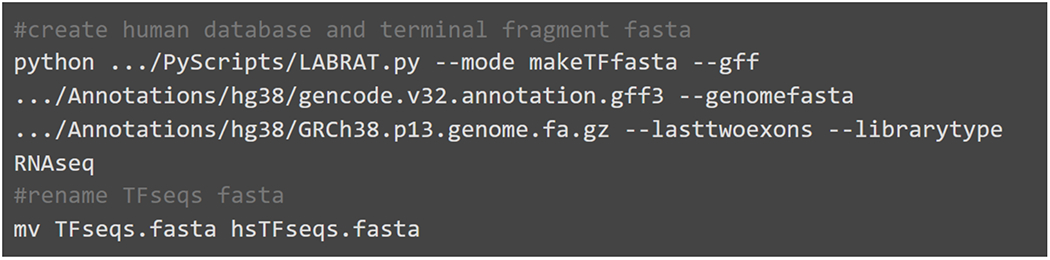
Example code for creating both the terminal fragment fasta file and a genome annotation database. Gencode’s genome annotation gff file and genome fasta are required inputs. TFseqs.fasta is created in the current directory while the database file is generated in the same directory as the gff.
