Fig. 4.
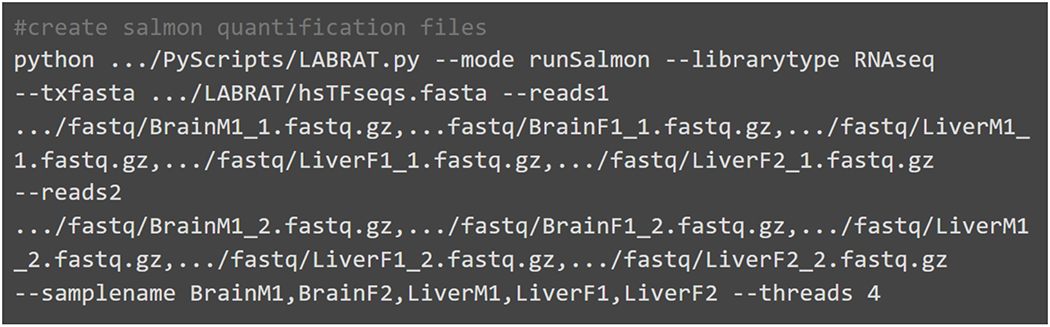
Example code for running LABRAT’s runSalmon function. RNAseq forward reads (reads1), reverse reads (reads2) and sample names are required inputs. Three prime end sequencing reads can also be used however the librarytype option should reflect the type of library provided. This code must be run in an empty directory as it outputs quantifications in new salmon directories for each sample.
