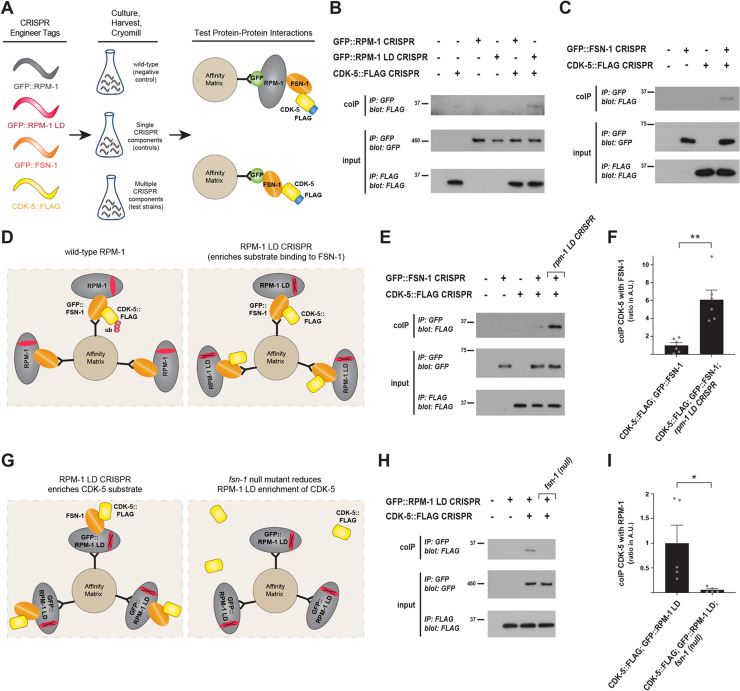
Fig 2. CRISPR-based biochemistry demonstrates substrate-like interactions between CDK-5 and the RPM-1/FSN-1 ubiquitin ligase complex.
A) Schematic of experimental workflow for CRISPR-based, native biochemistry using C. elegans. Multiple rounds of CRISPR/Cas9 engineering was used to affinity tag and point mutate components of the RPM-1/FSN-1 ubiquitin ligase complex and CDK-5. B) CoIP showing increased binding of CDK-5::FLAG to GFP::RPM-1 LD substrate ‘trap’ compared to GFP::RPM-1. C) CoIP showing CDK-5::FLAG binds GFP::FSN-1. D) Schematic showing how rpm-1 LD CRISPR substrate ‘trap’ enriches CDK-5 binding to FSN-1, if CDK-5 is an RPM-1/FSN-1 substrate. E) CoIP showing increased binding between CDK-5::FLAG and GFP::FSN-1 in rpm-1 LD CRISPR animals (far right lane) compared to wildtype background (second lane from right). F) Quantification of CDK-5 coIP with FSN-1 in rpm-1 LD CRISPR animals compared to wildtype background. G) Schematic showing how RPM-1 LD will not bind to CDK-5 in the absence of FSN-1 substrate recognition module (i.e. fsn-1 null mutant). H) CoIP showing reduced binding between CDK-5::FLAG and GFP::RPM-1 LD in fsn-1 (null) mutants (far right lane) compared to wildtype background (second lane from right). I) Quantification of CDK-5 coIP with RPM-1 LD in fsn-1 null mutants compared to wildtype background. For B, C, and E, representative of three or more independent experiments is shown. For H, representative of two independent experiments is shown. For F and I, shown are means from 5–6 replicates from 3 or 2 independent experiments, respectively. Wildtype values were normalized to 1 (Arbitrary Units) to facilitate comparisons. Error bars indicate SEM. Significance assessed using Student’s t-test. ** p<0.01.