(
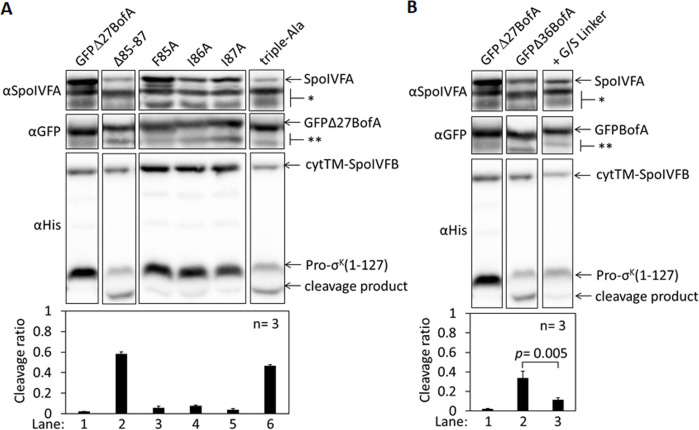
A) Cleavage assays examining the effects of a GFPΔ27BofA truncation and a triple-Ala substitution for the last three residues of GFPΔ27BofA. pET Quartet plasmids were used to produce Pro-σ
K(1–127), cytTM-SpoIVFB, SpoIVFA, and GFPΔ27BofA from pSO40 as a control (lane 1), GFPΔ27BofA lacking the last three residues (Δ85–87) from pSO43 (lane 2), or GFPΔ27BofA with a triple-Ala substitution for the last three residues from pSO67. Samples collected after 2 hr of IPTG induction were subjected to immunoblot analysis, and the graph shows quantification of the cleavage ratio, as explained in the
Figure 1B legend. For comparison, lanes 3–5 show data from
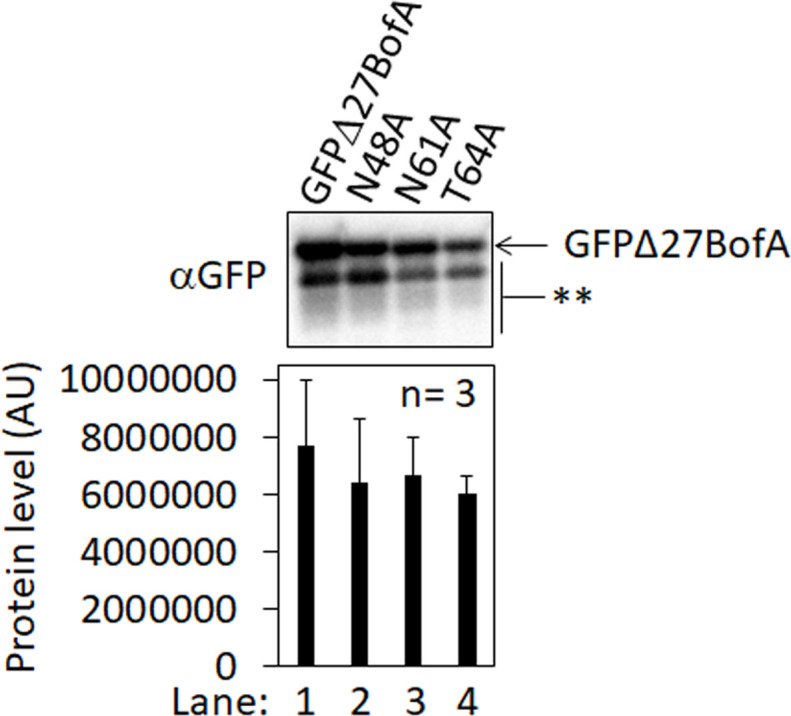
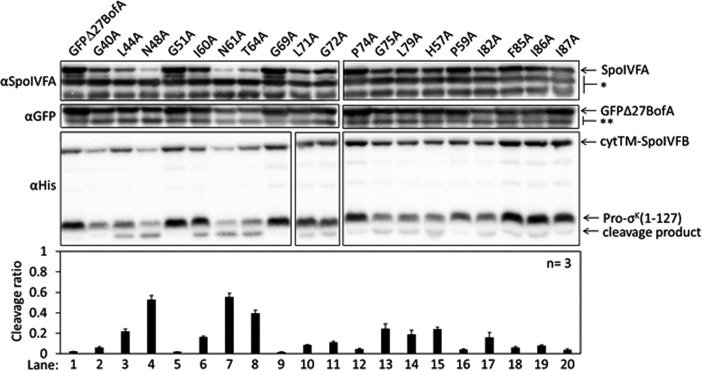
Figure 2 for single-Ala substitutions in GFPΔ27BofA. The triple-Ala variant increased the cleavage ratio (lane 6), as did the variant lacking the three residues (lane 2). Both variants accumulated normally, but less SpoIVFA accumulated compared to the controls, indicating that residues near the C-terminal end of GFPΔ27BofA affect the synthesis and/or stability of SpoIVFA, and contribute to inhibition of cytTM-SpoIVFB in
E. coli. (
B) GFPΔ27BofA lacks TMS1 and all but nine residues preceding predicted TMS2 (
Rudner and Losick, 2002). GFPΔ36BofA additionally lacks the nine residues. Cleavage assays were used to compare inhibition by GFPΔ27BofA, GFPΔ36BofA, or GFPΔ36BofA with a nine-residue glycine/serine (G/S) linker added between GFP and Δ36BofA. pET Quartet plasmids were used to produce Pro-σ
K(1–127), cytTM-SpoIVFB, SpoIVFA, and GFPΔ27BofA from pSO40 as a control lane 1, same data as in (
A), GFPΔ36BofA from pSO42 (lane 2), or GFPΔ36BofA with the nine-residue G/S linker from pSO69 (lane 3). Samples were subjected to immunoblot analysis and quantification as in (
A). Samples containing GFPΔ36BofA have a much greater cleavage ratio than samples containing GFPΔ27BofA. Since all four proteins accumulated well in both cases, the nine residues appeared to contribute to the inhibitory function of GFPΔ27BofA. Replacement of the nine residues with the G/S linker decreased the cleavage ratio, based on a Student’s two-tailed t-test (p value is indicated), suggesting that moving the GFP tag away from the membrane restored inhibitory function almost completely.