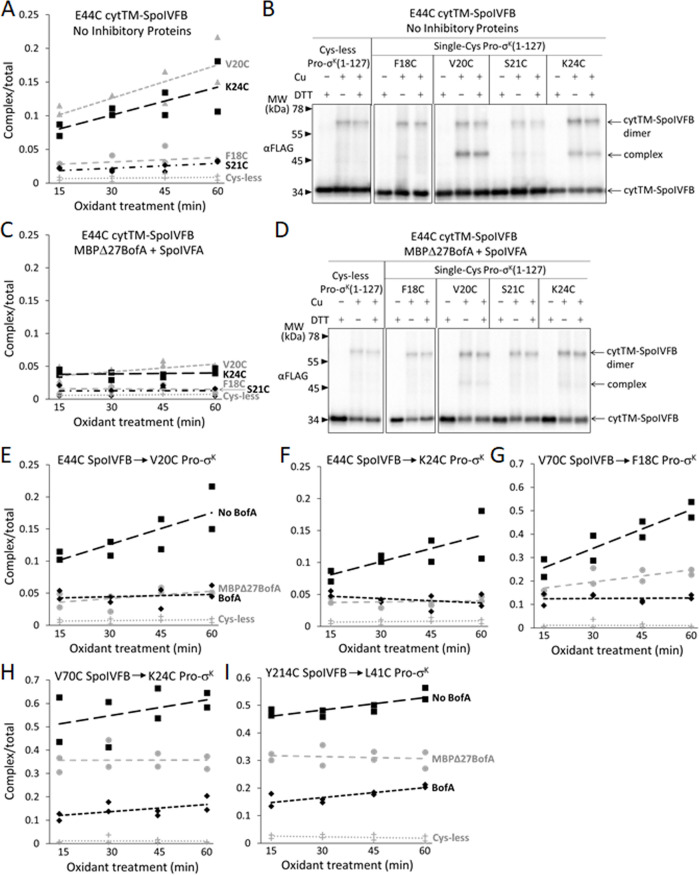
Figure 6. Inhibitory proteins block access of the Pro-σK(1–127) N-terminal region to the SpoIVFB active site and hinder interaction with the SpoIVFB membrane-reentrant loop and interdomain linker.
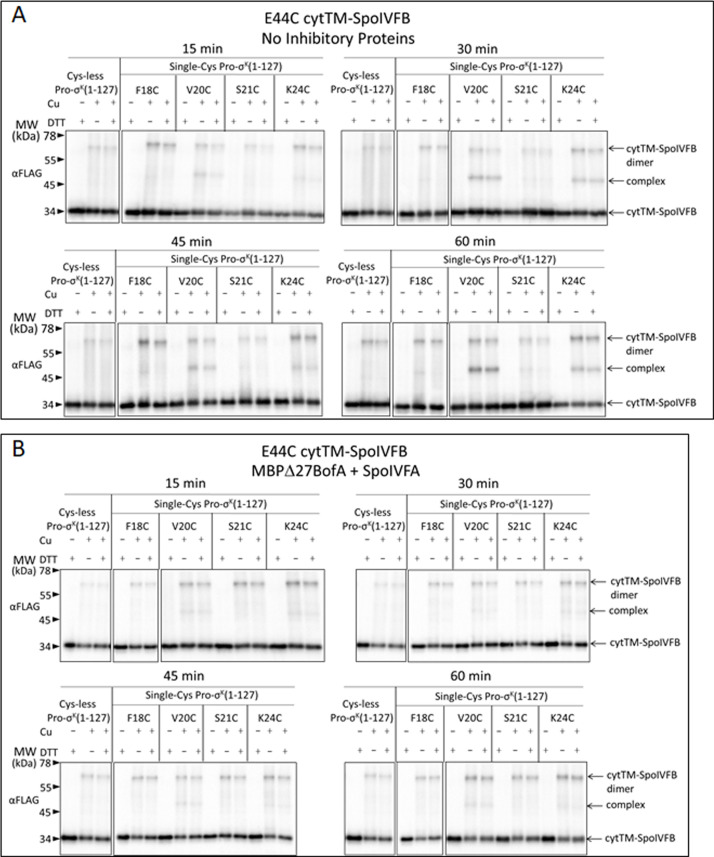
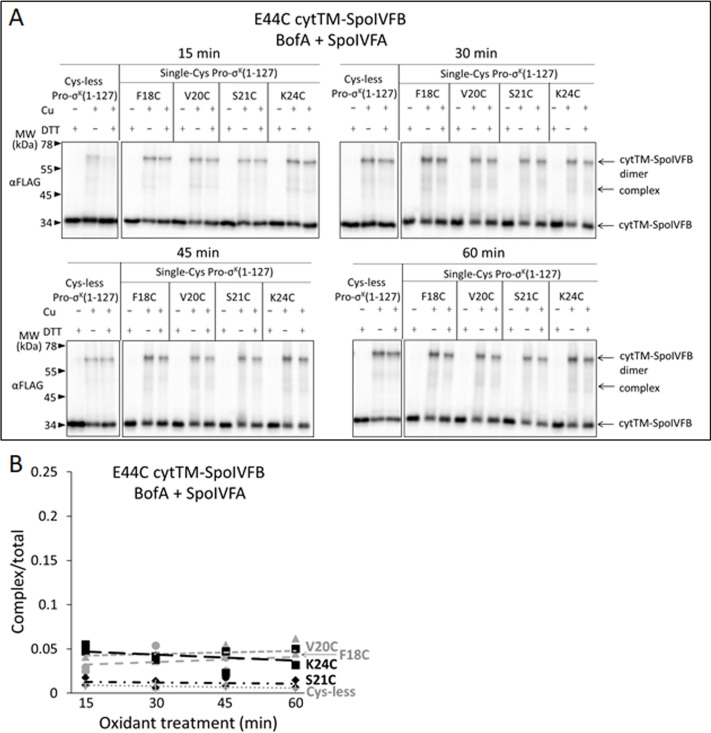
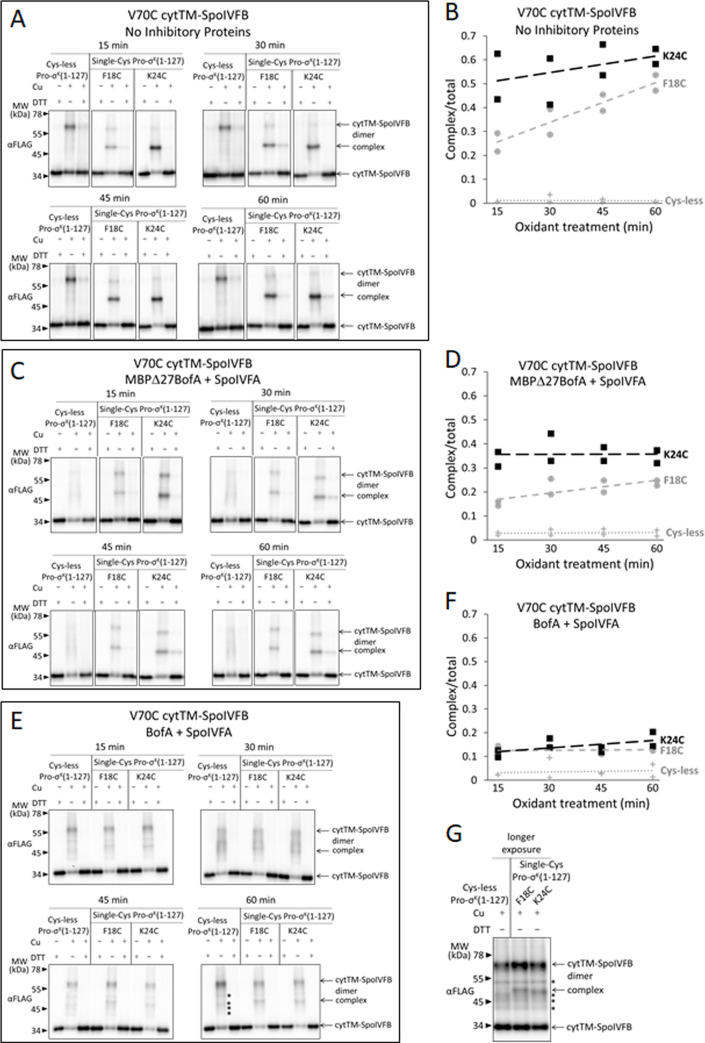
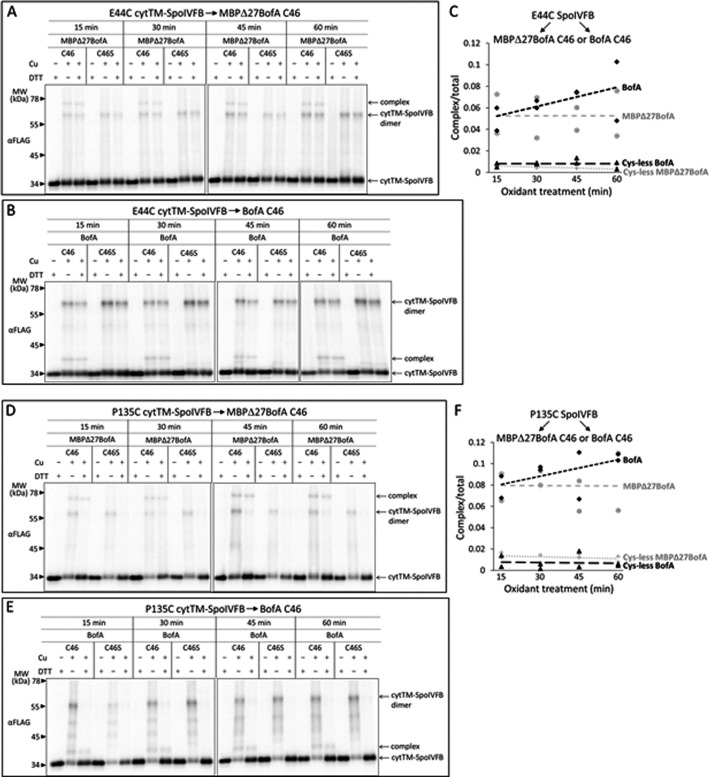
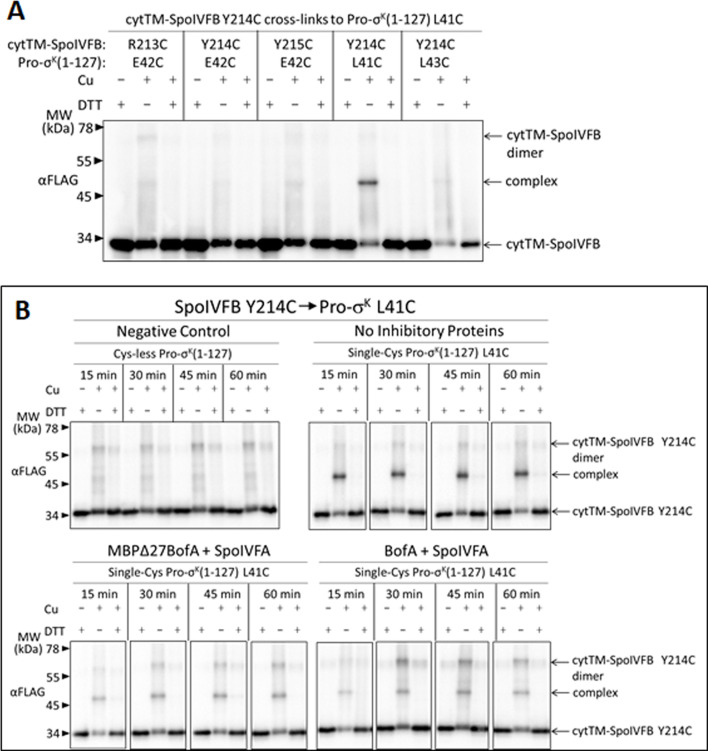
(A) E44C at the cytTM-SpoIVFB active site forms abundant disulfide cross-links with V20C or K24C in the N-terminal region of Pro-σK(1–127) in the absence of inhibitory proteins. pET Duet plasmids were used to produce single-Cys E44C cytTM-SpoIVFB in combination with single-Cys F18C (pSO167), V20C (pSO169), S21C (pSO170), or K24C (pSO128) Pro-σK(1–127), or with Cys-less Pro-σK(1–127) (pSO79) as a negative control, in Escherichia coli. Samples collected after 2 hr of IPTG induction were treated with Cu2+(phenanthroline)3 oxidant for 15, 30, 45, or 60 min to promote disulfide bond formation and subjected to immunoblot analysis with FLAG antibodies to visualize the cytTM-SpoIVFB monomer, dimer, and complex with Pro-σK(1–127) (Figure 6—figure supplement 1A). Abundance of the complex was divided by the total amount of cytTM-SpoIVFB monomer, dimer, and complex. The ratio over time was plotted (n=2) with a best-fit trend line. (B) Representative immunoblots of 60 min samples from the experiment are described in (A). (C) MBPΔ27BofA and SpoIVFA decrease cross-linking between E44C cytTM-SpoIVFB and V20C or K24C Pro-σK(1–127). pET Quartet plasmids were used to produce single-Cys E44C cytTM-SpoIVFB in combination with single-Cys F18C (pSO163), V20C (pSO165), S21C (pSO166), or K24C (pSO131) Pro-σK(1–127), or with Cys-less Pro-σK(1–127) (pSO110) as a negative control, and Cys-less variants of MBPΔ27BofA and SpoIVFA in E. coli. Samples collected after 2 hr of IPTG induction were treated and subjected to immunoblot analysis as in (A) (Figure 6—figure supplement 1B). The complex/total ratio was plotted as in (A). (D) Representative immunoblots of 60 min samples from the experiment are described in (C). (E, F) Summaries of the effects of inhibitory proteins on cross-linking between E44C cytTM-SpoIVFB and V20C or K24C Pro-σK(1–127). Data from (A) (labeled ‘No BofA’ in (E), although SpoIVFA is also absent), (C) (labeled ‘MBPΔ27BofA’ in (E), although SpoIVFA is also present), and Figure 6—figure supplement 2 (labeled ‘BofA’ in (E), although SpoIVFA is also present) are plotted along with Cys-less Pro-σK(1–127) as a negative control. In (F), symbols and lines are as in (E). (G, H) Summaries of the effects of inhibitory proteins on cross-linking between V70C in the cytTM-SpoIVFB membrane-reentrant loop and F18C or K24C in the Pro-σK(1–127) N-terminal region. Data from Figure 6—figure supplement 3 are plotted using symbols and lines as in (E). (I) Summary of the effects of inhibitory proteins on cross-linking between Y214C in the cytTM-SpoIVFB interdomain linker and L41C in the Pro-σK(1–127) N-terminal region. Data from Figure 6—figure supplement 5 are plotted using symbols and lines as in (E).