Figure 1. Ankmy2 Knockout Mice Are Embryonic Lethal at 10 to 12-Somite Stage and Exhibit Completely Open Neural Tubes.
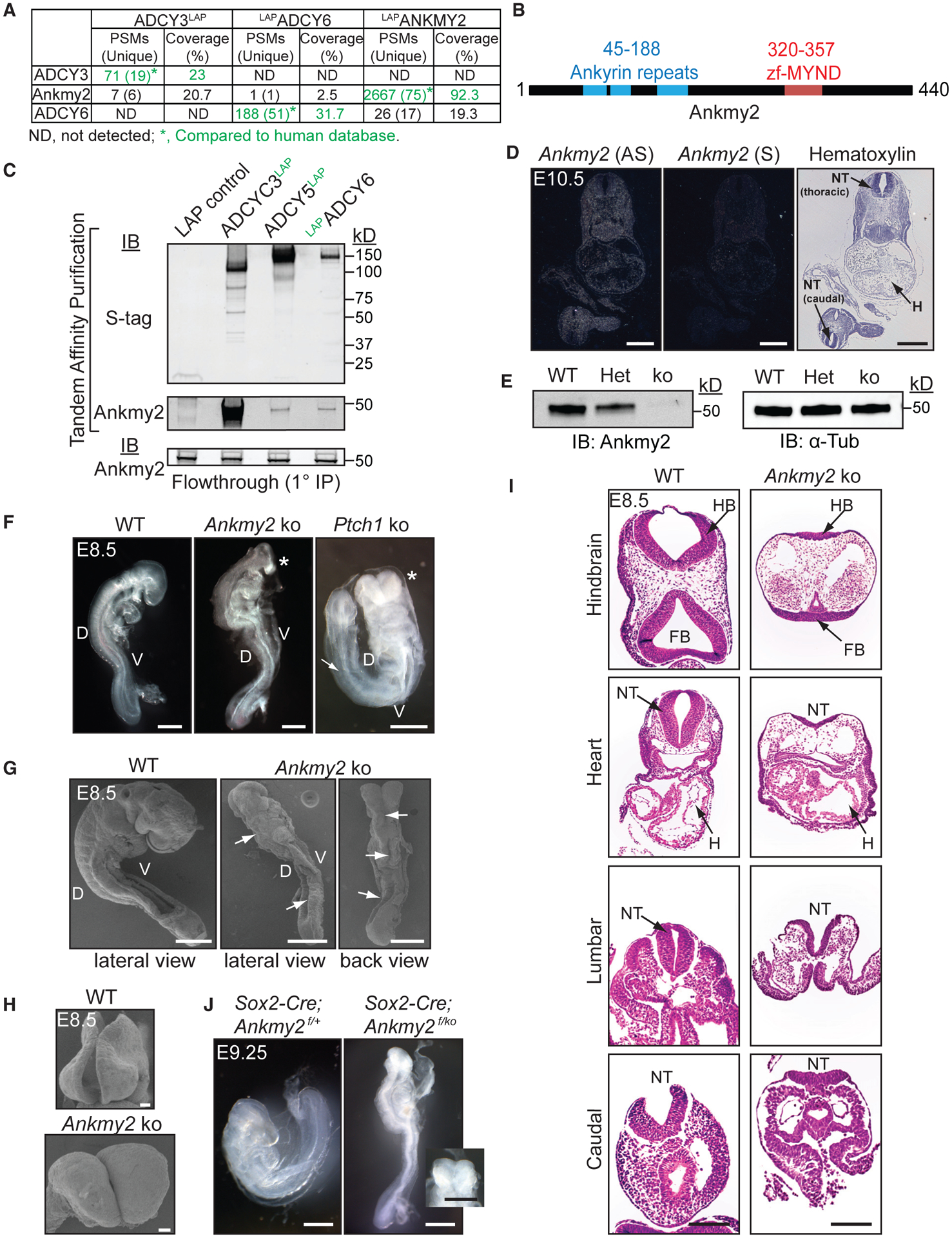
(A) TAP-MS of LAP-tagged ACs or ANKMY2 stably expressed in NIH 3T3 cells. ADCY3/6 and ANKMY2 clones were human ORFs, and peptides were compared to the human database.
(B) Ankmy2 protein domains. zf-MYND, zinc finger MYND domain.
(C) TAP of ADCY3/5/6 detects Ankmy2. Flow through (0.35%) from the initial anti-GFP IP below. n = 4 independent experiments.
(D) Radioisotopic RNA in situ hybridization using antisense or sense (AS or S) riboprobes for Ankmy2 in transverse sections of wild-type E10.5 embryos.
(E) Immunoblotting of whole-embryo lysates at E8.5 in wild-type (WT), heterozygote (Het), and Ankmy2 knockout (ko). α-tubulin, loading control. n = 3 embryos per genotype.
(F) Bright-field images of WT (16 somites), Ankmy2 ko (10–12 somites), and Ptch1 ko (14–16 somites) whole-mount embryos dissected at E8.5. Asterisks, rostral malformations. n > 10 (WT, Ankmy2 ko), 6 (Ptch1 ko).
(G and H) Scanning electron micrographs of WT (12–14 somites) (G) and Ankmy2 ko (12 somites) and en face views of head regions (H). Arrows, open neural tube. n = 3 (WT), 5 (ko).
(I) HE-stained transverse sections of WT (16 somites) and Ankmy2 ko (12 somites). n = 4 per genotype.
(J) Sox2-Cre; Ankmy2f/ko (12 somites) embryos dissected at E9.25 recapitulates Ankmy2 ko compared with control (20–22 somites) (inset: en face head view). n = 2 per genotype.
Scale: D, 500 μm; F and G, I and J (including inset), 200 μm; H, 20 μm.
Abbreviations: PSM, peptide-to-spectral matches; D, Dorsal; V, Ventral; HB, hindbrain; FB, Forebrain; H, Heart; NT, neural tube; WT, wild-type.
