Abstract
Coronavirus disease 2019 (COVID-19) is a potentially life-threatening infection characterized by excessive inflammation, coagulation disorders and organ damage. A dysregulated myeloid cell compartment is one of the most striking immunopathologic signatures of this newly emerged infection. A growing number of studies are reporting on the expansion of myeloid cells with immunoregulatory activities in the periphery and airways of COVID-19 patients. These cells share phenotypic and functional similarities with myeloid-derived suppressor cells (MDSCs), which were first described in cancer patients. MDSCs are a heterogeneous population of pathologically activated myeloid cells that exert immunosuppressive activities against mainly effector T cells. The increased frequency of these cells in COVID-19 patients suggests that they are involved in immune regulation during this infection. In this article, we review the current findings on MDSCs in COVID-19 and discuss the complex role of these cells in the immunopathology of COVID-19.
Keywords: Myeloid-derived suppressor cell, SARS-CoV-2, COVID-19, Immunopathology
Graphical abstract
1. Introduction
Massive expansion of the myeloid cell compartment and a decrease in the leukocyte compartment have been repeatedly observed in patients with severe COVID-19, emphasizing an important role of myeloid cells in the pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection [[1], [2], [3]]. Myeloid cells in COVID-19 are characterized by diminished antigen-presenting and increased immunosuppressive characteristics, both of which are consistent with the MDSC profile [4,5]. MDSCs are pathologically activated monocytes and granulocytes with unique metabolic and gene expression profiles that attract special attention due to their ability to induce systemic and local immunosuppression and promote accumulation of other immunosuppressive cells, such as T-regulatory cells [6]. The negative role of this cell population was first established in tumor immunology. In cancer settings, these cells have been shown to exhibit strong antigen-specific and antigen-nonspecific suppression of CD4+ and CD8+ T cells, as well as some populations of innate immune cells, promoting tumor growth and exacerbating disease [6].
Later, MDSCs were described in a number of viral diseases, including some respiratory viral infections [7]. An accumulation of MDSCs was detected in the lungs of mice with influenza A virus (IAV) infection and in the peripheral blood of patients with IAV [8,9]. These cells have been shown to suppress T cells via production of inducible NO synthase (iNOS) and arginase 1(Arg1) [8]. The role of these cells in the pathogenesis of infectious diseases remains obscure. On one hand, MDSCs suppress functions of effector immune cells, which may result in the inability of the organism to develop an efficient immune response against microbes and hinder the elimination of pathogens from the bloodstream and the site of infection. On the other hand, MDSCs can limit the hyperinflammation and “cytokine storm” triggered by infection and protect host organs from fatal dysfunction [10]. In this article, we provide a comprehensive compilation of the current knowledge about MDSCs in COVID-19 and discuss a possible role of these cells in the immunopathogenesis of SARS-CoV-2 infection.
2. Overview of COVID-19 pathogenesis
SARS-CoV-2 is a new coronavirus that emerged in China in December 2019 and spread rapidly across the world causing more than 5 million reported deaths worldwide within two years [11]. SARS-CoV-2 utilizes the host angiotensin-converting enzyme 2 (ACE2) receptor for cell entry into a number of organs [12]. Infection begins with recognition of the receptor-binding domain (RBD) of the S protein of SARS-CoV-2 by the ACE2 receptor on epithelial cells of the upper respiratory tract and subsequent intracellular protease–mediated proteolytic cleavage of the S protein, which allows entry of the virus [13]. The respiratory system is a major target of the virus, but single-cell RNA sequence studies of ACE2 receptor expression and necropsy studies indicate that the virus is also capable of invading the kidneys, pancreas, small intestine, heart, and blood vessels, which may explain a wide range of non-respiratory complications observed in COVID-19 patients [14,15].
Replication of SARS-CoV-2 in host cells leads to an accumulation of virus-derived pathogen-associated molecular patterns (PAMPs), which in turn are recognized by a number of host membrane or cytosolic pattern recognition receptors (PRRs), such as Toll-like receptor 3 (TLR3), TLR4, TLR7, TLR8, NOD-like receptor family porin domain containing 3 (NLRP3), retinoic acid–inducible gene 1 (RIG1), melanoma differentiation-associated protein 5 (MDA5), and LGP2 [[16], [17], [18]]. TLR7 and TLR8 recognize the single-stranded RNA of SARS-CoV-2, while TLR3, RIG1, LGP2, and MDA5 bind double-stranded RNA intermediates generated during viral replication [[17], [18], [19], [20]]. TLR4 and NLRP3 recognize SARS-CoV-2 proteins [21,22]. Viral replication also leads to the expression of host-derived danger-associated molecular patterns (DAMPs), which are then released extracellularly from injured or dying infected cells upon rupture of the plasma membrane to be recognized by PRR-bearing cells. Major DAMPs associated with COVID-19 include calprotectin S100A8/A9, high mobility group box 1 protein (HMGB1), mitochondrial DNA (MT-DNA), and extracellularly-secreted nicotinamide phosphoribosyl-transferase (eNAMPT) [[23], [24], [25]].
Recognition of PAMPs and DAMPs by PRRs and other innate receptors triggers complicated downstream signaling pathways leading to the first wave of pro-inflammatory mediators, chemokines and growth factors, including interleukin 1 (IL-1), IL-6, interferon α/β (IFNα/β), tumor necrosis factor α (TNFα), C-X-C motif chemokine ligand 8 (CXCL8/IL-8), CXCL10, C—C motif chemokine ligand 5 (CCL5), granulocyte colony-stimulating factor (G-CSF), and granulocyte-macrophage colony-stimulating factor (GM-CSF), produced by type I and type II alveolar cells, alveolar macrophages, resident innate lymphoid, epithelial, and endothelial cells [[26], [27], [28], [29]]. Production of these pro-inflammatory mediators triggers an innate immune response that increases vascular permeability and recruits PRR-expressing myeloid cells such as neutrophils, monocytes, dendritic cells, as well as platelets and NK cells to the lungs. Once in the lungs, these immune cells are in turn activated to produce a second wave of cytokines and chemokines as well as cytotoxic mediators such as reactive oxygen species, proteases, and granular enzymes, aimed at limiting viral amplification within infected cells [27]. The innate immune response also primes the adaptive immune response, which gets activated over the course of several days [29]. The adaptive immune response is orchestrated by antigen-specific B cells, CD4+ T helper and CD8+ cytotoxic T cells. CD4+ T cell responses against SARS-CoV-2 have been reported more frequently than CD8+ T cell responses [30]. Seroconversion in individuals with SARS-CoV-2 infection is usually observed 5–15 days after the onset of symptoms [31,32]. Therefore, first the innate and later the adaptive immune system act synchronously to eliminate the virus and the cells damaged by the virus. After pathogen elimination, a number of immunoregulatory cell populations resolve the inflammatory response and restore tissue homeostasis. The major immunoregulatory cell subsets that contribute to the resolution of inflammation are M2 macrophages, regulatory dendritic cells, T-regulatory cells and MDSCs [10].
While the majority of patients recover within 2 weeks after symptom onset, approximately 14% of patients develop severe or critical illness, often requiring hospitalization and ventilation support [33]. Underlying medical conditions and advanced age increase the risk of hospitalization sixfold [33]. The medical conditions that increase the risk of severe manifestations and death from COVID-19 include hypertension, diabetes, cancer, and chronic respiratory, cerebrovascular, and renal diseases [34]. This risk is multifactorial. On one hand, the above-mentioned comorbidities, as well as advanced age, are associated with an immunocompromised state characterized by dysregulated immune responses and thus a weakened ability to fight the virus [29,[35], [36], [37]]. On the other hand, the virus itself has evolved mechanisms to suppress antiviral immune responses. The type I and type III IFN systems are critical for early antiviral innate immune responses [[38], [39], [40]]. SARS-CoV-2 is able to successfully impede IFN responses by several mechanisms [[40], [41], [42], [43]]. Disruption of IFN production allows rapid replication of the virus, resulting in high viral loads early in the disease. High viral loads and/or impaired immune responses can trigger an overproduction of pro-inflammatory cytokines known as the “cytokine storm”, leading to life-threatening systemic inflammation, thromboembolic disorders, and multiple organ failure [44]. Specifically in the respiratory tract, viral destruction of alveolar cells and bronchial epithelial cells, along with mass production of cytokines, leads to pneumonia and, in severe cases, acute respiratory distress syndrome (ARDS) [45,46]. Although the full picture of COVID-19 pathogenesis remains to be investigated, it is now clear that the cytokine and chemokine profile observed in COVID-19 patients promotes profound changes in myeloid cells that appear to be able to determine the outcome of infection [5,47].
3. Main characteristics and functions of MDSCs
The immunoregulatory role of MDSCs has been described in a number of pathological (cancer, sepsis, chronic inflammation, autoimmune diseases, infections, obesity) and physiological conditions (pregnancy, aging) [48]. Two main groups of MDSCs have been described in both humans and mice, namely granulocytic or polymorphonuclear MDSCs (PMN-MDSCs) and monocytic MDSCs (M-MDSCs). In humans, PMN-MDSCs are most commonly described as CD11b+CD14−CD15+/CD66b+, whereas M-MDSCs are mainly defined as CD11b+HLA-DRdim/-CD14+CD15−. Both cell subpopulations also express the myeloid marker CD33 [49]. Another subpopulation of MDSCs that has been described in humans is called early-stage MDSCs (e-MDSCs). Early-stage MDSCs are immature myeloid progenitor cells with suppressive properties defined as Lin(including CD3, CD14, CD15, CD19, CD56) −HLA-DR−CD33+ [50]. Phenotypic identification of PMN-MDSCs, M-MDSCs and eMDSCs is challenging as they share similar cell surface phenotypes with classical neutrophils, monocytes and basophils, correspondingly [50,51]. A number of approaches have been described to differentiate MDSC subpopulations from the effector immune cell subpopulations. PMN-MDSCs from peripheral blood are enriched on lower density gradients of 1.077 g/ml, whereas classical neutrophils are separated on higher density gradients [50]. Using whole-genome analysis and single-cell RNA sequencing, several cell surface markers expressed by human MDSCs in cancer patients have been identified, including lectin-type oxidized LDL receptor 1 (LOX1), programmed death ligand 1 (PDL1), CD84, and CXCR1 [[52], [53], [54]]. MDSC subpopulations share similar biochemical and molecular parameters that warrant their immunosuppressive potential, including upregulation of signal transducer and activator of transcription 3 (STAT3) expression and genes responsible for Arg1 and S100A8/A9 expression [6].
Phenotypic identification of MDSCs should be followed by an assessment of their ability to inhibit activity of effector immune cells. Although MDSCs can suppress various cells of the immune system, including NK cells and B cells, their main function is to induce T cell tolerance [[55], [56], [57]]. PMN-MDSCs mainly induce antigen-specific T cell tolerance via the production of reactive oxygen species (ROS), peroxynitrite (PNT), Arg1, and prostaglandin E2 (PGE2), whereas M-MDSCs induce antigen-specific and nonspecific T cell tolerance via the production of nitric oxide (NO), indoleamine 2,3-dioxygenase (IDO), immunosuppressive cytokines such as IL-10 and tumor growth factor β (TGFβ), and expression of PDL1 [48]. MDSC-derived ROS and PNT, which is the product of the reaction of ROS and NO, induce nitration of the T cell receptor on CD8+ cells during cell-to-cell contacts [58]. This modification causes T cells to lose their ability to bind the phosphorylated major histocompatibility complex (MHC) and therefore fail to respond to specific antigens, resulting in antigen-specific T cell tolerance [59]. Arg1 is responsible for degradation of the non-essential amino acid L-arginine. The degradation of L-arginine blocks the proliferation of T cells and leads to molecular changes in T cells, including low expression of the CD3ζ chain and decreased production of IFNγ [60]. IL-10 and TGFβ produced by MDSCs inhibit T cell activation and recruit T regulatory cells [61]. Moreover, MDSCs can induce apoptosis of T cells through cell-to-cell contacts by binding PD1 molecules on T cells with their ligand PDL1 [62].
According to an experimental algorithm for reporting MDSCs proposed by Bronte et al. in 2016, biochemical and molecular characteristics of myeloid cells should be assessed in case of inability to demonstrate their direct antigen-specific or antigen-nonspecific suppressive activity towards T cells [50]. Up-regulated immunosuppressive molecules and alterations in MDSC transcription factors and regulators (such as interferon regulatory factor 8, STAT3, and S100A8/A9) allows characterization of myeloid cells as MDSC-like cells (MDSC-LC). However, considering that functional and molecular assays on human MDSCs are complicated to perform in routine laboratory practice due to the difficulties in MDSC isolation and that this may lead to substantial underreporting of MDSCs in COVID-19, we discuss here not only studies that identified MDSCs and MDSC-LCs but also studies that identified myeloid cell subsets with the MDSC phenotype without further functional, molecular, or biochemical characterization. Therefore, caution should be exercised in further interpretation of these data.
4. MDSCs in COVID-19
4.1. Phenotype and sites of detection
MDSCs have been analyzed mainly in the peripheral blood of COVID-19 patients. Several studies have reported the frequency of MDSCs in the airways of COVID-19 patients. A study by Falck-Jones et al. analyzed the distribution of M-MDSCs in nasopharyngeal (NPA) and endotracheal (ETA) aspirates from mild, moderate, and severe patients [63]. They showed that the frequency of M-MDSCs was not increased in either NPA or ETA in COVID-19 patients compared with healthy donors. A study by Dean et al. reported large aggregations of Arg1-expressing PMN-MDSCs in the lungs of patients who died from COVID-19 [64]. These data suggest that although SARS-CoV-2 infection starts in the upper respiratory tract, it progresses in the lower parts of airways, where local recruitment of MDSCs is observed.
All studies we identified reported an increase in either the proportions or counts of myeloid cells with the MDSC phenotype in the peripheral blood of COVID-19 patients, particularly with moderate and severe infection compared with healthy donors (Table 1 ). A few studies used LOX1 as a specific marker for PMN-MDSCs [63,65,66]. Most importantly, several studies reported the direct suppressive activity of these cells as estimated in in vitro experiments (Table 1). The frequency of MDSCs was reported as high as 90% of the total circulating mononuclear cells in patients with severe disease and up to 25% in patients with mild or moderate disease [67]. Moreover, MDSCs levels have been shown to remain elevated after recovery from mild and even asymptomatic COVID-19 [68], likely contributing to the recently described post-COVID-19 immunodepression [69].
Table 1.
Summary of current studies on MDSCs in COVID-19 patients.
| # | Study size (COVID-19 patients + Healthy donors (HD)) | Source of MDSC | Density gradient centrifugation | Fresh/cryopreserved samples | Markers of MDSCs | MDSC frequency | Functional assay in vitro | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | N = 180 (COVID-19: 19 mild, 58 moderate, 58 severe, 12 fatal; HD: 33) | Peripheral blood (PB), nasopharyngeal (NPA) and endotracheal (ETA)aspirates | Yes | Fresh | M-MDSC: Lin−HLA-DR−CD14+ PMN-MDSC: Lin−HLA-DR−CD66abce+LOX1+ |
Increase in M-MDSC and PMN-MDSC in PB (proportion, in COVID-19 patients as compared to HD, in severe patients as compared to mild and moderate patients) No change in M-MDSC and PMN-MDSC in NPA and ETA (proportion) |
Inhibition of antigen-nonspecific T cell proliferation and IFNγ production | [63] |
| 2 | N = 40 (COVID-19: 20 mild; HD: 20) | PB | No | Fresh | M-MDSC: CD11b+HLA-DR−CD33+ CD14+CD15− PMN-MDSC: CD11b+HLA-DR−CD33+CD14−CD15+ |
Increase in M-MDSC (counts, in COVID-19 patients as compared to HD) No change in PMN-MDSC (counts, in COVID-19 patients as compared to HD) |
None | [73] |
| 3 | N = 59 (COVID-19: 38 acute, 10 recovered; HD: 11) | PB | Yes | Cryopreserved | M-MDSC: Lin−HLA-DR−CD33+CD14+CD15− PMN-MDSC: CD15+CD16+LOX1+ |
Increase in M-MDSC and PMN-MDSC (proportion, in acute COVID-19 patients as compared to HD) | None | [65] |
| 4 |
N = 70 (COVID-19: 5 asymptomatic, 24 severe, 10 deceased, 26 convalescent; HD: 15) |
PB, lungs | Yes | Fresh | M-MDSC: HLA-DR-/dimCD14+ PMN-MDSC: CD66b+HLA-DR−CD14− |
Increase in M-MDSC and PMN-MDSC in PB (proportion, in severe patients as compared to convalescent, asymptomatic and HD) PMN-MDSC infiltrates in the lungs |
None | [64] |
| 5 | N = 158 (COVID-19: 32 non-ICU, 96 ICU; HD: 30) | PB | Yes | Fresh | PMN-MDSC: Lin−HLA-DR−CD11b+CD33+CD15+CD14− | Increase in PMN-MDSC (proportion, in COVID-19 patients as compared to HD, in ICU patients as compared to non-ICU) | Inhibition of antigen-nonspecific proliferation and antigen-specific IFNγ production by T cells | [71] |
| 6 |
N = 68 (COVID-19: 41 non-ICU; 7 ICU; HD: 20) |
PB | Yes | Fresh | Customized Duraclon tubes | Increase in MDSC (proportion, in COVID-19 patients as compared to HD) | None | [85] |
| 7 | N = 26 (COVID-19: 9 mild, 9 severe; HD: 8) | PB | Yes | Fresh | PMN-MDSC: Lin−HLA-DRdim/−CD11b+CD33+CD15+CD14− | Increase in PMN-MDSC (proportion, in COVID-19 patients as compared to HD, in severe patients as compared to mild patients) | Inhibition of antigen-nonspecific T cell proliferation and IFNγ and TNFα production |
[67] |
| 8 | N = 39 (COVID-19: 13 moderate, 13 severe; HD: 13) | PB | Yes | Fresh | M-MDSC: HLA-DRdimCD14+; PMN-MDSC: Lin−HLA-DRdimCD16+CD15+ e-MDSC: Lin−HLA-DRdimCD33+CD15+ |
Increase in M-MDSC and PMN-MDSC (count, in COVID-19 patients as compared to HD, in severe patients as compared to moderate patients) | No | [75] |
| 9 | N = 57 (COVID-19: 21 mild, 20 severe; HD: 16) | PB | Yes | Fresh | M-MDSC: Lin−HLA-DRdim CD11b+CD14+; PMN-MDSC: Lin−HLA-DRdimCD11b+CD15/CD66b+ e-MDSC: Lin−HLA-DRdimCD66b/CD15−CD14− |
Increase in M-MDSC and PMN-MDSC (proportion and count, in severe COVID-19 patients as compared to HD and mild patients) | Inhibition of antigen-nonspecific T cell proliferation | [76] |
| 10 | N = 47 (COVID-19: 12 mild,15 moderate, 13 severe; HD: 7) | PB | Yes | Cryopreserved | M-MDSC: Lin−HLA-DR−CD11b+CD33+CD14+CD15−; PMN-MDSC: Lin−HLA-DR−CD11b+CD33+CD14−CD15+ e-MDSC: Lin−HLA-DR−CD11b+CD33+CD14−CD15− |
Increase in PMN-MDSC (proportion, in severe discharged patients as compared to HD, mild, moderate, and severe deceased patients). No change in M-MDSc and e-MDSC (proportion, in patients as compared to HD) |
No | [72] |
| 11 | N = 61 (COVID-19: 39 ICU; HD: 22) | PB | No | Fresh | PMN-MDSC: CRTH2− CD15+LOX-1+CD10dimCD16dim | Increase in PMN-MDSC (proportion, in ICU patients as compared to HD) | Inhibition of antigen-nonspecific T cell production of IFN-g |
[66] |
| 12 | N = 27 (COVID-19: 19 non-ICU; 8 ICU; HD: 11) | PB | No | Fresh | M-MDSC: HLA-DR-/dimCD14+ | Increase in M-MDSC (proportion, in ICU patients as compared to non-ICU and HD, in non-ICU as compared to HD) | No | [86] |
| 13 | N = 57 (COVID-19: 21 mild, 15 severe, 9 convalescent; HD: 12) | PB | Yes | Fresh | M-MDSC: Lin−HLA-DR-/dimCD11b+CD33+CD14+CD15− PMN-MDSC: Lin−HLA-DR-/dimCD11b+CD33+CD14−CD15+ e-MDSC: Lin−HLA-DR−CD11b+CD33+CD14−CD15− |
Increase in M-MDSC and PMN-MDSC (proportion, in severe patients as compared to mild, convalescent patients and HD). No change in e-MDSC (proportion) |
No | [2] |
| 14 | N = 58 (COVID-19: 13 moderate, 37 severe; HD: 8) | PB | No | Fresh | M-MDSC: HLA-DR-/dimCD11b+CD14+CD15− PMN-MDSC: HLA-DR-/dimCD11b+CD14−CD15+ |
Increase in M-MDSC and PMN-MDSC (proportion, in patients as compared to HD) | No | [84] |
| 15 | N = 26 (COVID-19: 13 convalescent; HD: 13) | PB | Yes | Fresh | M-MDSC: Lin−HLA-DR-/dimCD11b+CD14+CD15− PMN-MDSC: Lin−HLA-DR-/dimCD11b+CD14−CD15+ e-MDSC: Lin−HLA-DR−CD11b+ CD14−CD15− |
Increase in PMN-MDSC (proportion, in convalescent patients as compared to HD). No change in M-MDSC and e-MDSC (proportion) |
Inhibition of antigen-nonspecific T cell proliferation | [68] |
| 16 | N = 67 (COVID-19: 26 mild, 15 severe; HD: 26) | PB | Yes | Fresh | M-MDSC: HLA-DR−CD11b+CD66b−CD14+CD15− PMN-MDSC: HLA-DR−CD11b+CD66b+CD14−CD15+ |
Increase in M-MDSC and PMN-MDSC (proportion, in patients as compared to HD; increase in PMN-MDSC in mild patients as compared to severe patients) | No | [70] |
| 17 | N = 120 (COVID-19: 27 mild, 47 severe, 46 deceased) | PB | No | Fresh | M-MDSC: HLA-DR−CD14+ |
Increase in M-MDSC (proportion, in deceased patients as compared to mild and severe patients) | No | [3] |
| 18 | N = 80 (COVID-19: 40 ICU discharged, 40 ICU deceased) | PB | No | Fresh | M-MDSC: HLA-DR-/dimCD11b+CD33+CD14+CD15− PMN-MDSC: HLA-DR-/dimCD11b+CD33+CD14−CD15+ |
Increase in PMN-MDSC (counts, in ICU deceased patients as compared to ICU discharged) | No | [103] |
| 19 | N = 20 (COVID-19: 11 non-ICU, 9 ICU) | PB | Yes | Cryopreserved | M-MDSC: Lin−HLA-DR−CD11b+CD33+CD14+CD15− PMN-MDSC: Lin−HLA-DR−CD11b+CD33+CD14−CD15+ |
Increase in total MDSC (proportion, in ICU patients as compared to non-ICU) | No | [105] |
Intriguingly, while some studies reported a substantial increase in both circulating M-MDSC and PMN-MDSC subpopulations [2,63,64,70], other studies failed to identify M-MDSCs in either mild, moderate, or severe COVID-19 patients at different observation time points [67,71]. Takano et al. found an elevated frequency of PMN-MDSCs but no M-MDSCs in severe COVID-19 [72]. In contrast, a study by Jiménez-Cortegana et al. found a fourfold increase in myeloid cells with the M-MDSC phenotype in COVID-19 patients, but not in myeloid cells with the PMN-MDSC phenotype [73]. Overall, increased numbers of PMN-MDSCs in COVID-19 patients have been reported more frequently than M-MDSCs (Table 1). These variations may be explained by differences in experimental design, including different approaches to sample handling (use of whole blood samples versus peripheral blood mononuclear cell fractions, use of thawed versus freshly isolated samples) and phenotypic identification of MDSC subpopulations, which varied across studies (Table 1), as well as immunomodulatory effects of drugs received by patients. However, it is tempting to speculate that another reason could be different cytokine or chemokine profiles favoring the development and expansion of specific MDSC subpopulations in COVID-19 patients due to different clinical courses of the disease. For example, it was recently suggested that recruitment of M-MDSC and PMN-MDSC subpopulations might be driven by different chemokines [74].
There was no evidence of increased numbers of e-MDSCs in the peripheral blood of COVID-19 patients [2,68,72,75,76].
Single-cell RNA sequencing studies identified molecular and metabolic signatures of MDSCs in COVID-19 patients [65]. Monocytic cells from patients with severe SARS-CoV-2 infection repeatedly show increased expression of genes encoding calprotectin (S100A8/A9) and calgranulin C (S100A12), in parallel with reduced expression of genes encoding the class II MHC (MHC-II) [4,5,65,[77], [78], [79], [80]]. PMN-MDSCs from severe COVID-19 patients are characterized by upregulated expression of genes associated with immunosuppressive functions, including Arg1 and iNOS, genes encoding S100A8/A9 and S100A12, and genes involved in the IL-1 signaling and NF-κB activation pathways [4,5,64]. A single-cell RNA sequencing study by Thompson et al. identified a unique population of PMN-MDSCs that expresses both voltage-dependent anion channel 1 (VDAC1) and the initial enzyme of glycolysis, hexokinase II (HKII), in acute COVID-19 patients but not in recovered patients or patients with other viral diseases (influenza, hepatitis C). They also found that M-MDSCs from COVID-19 patients expressed high levels of the VDAC1 and carnitine palmitoyltransferase 1a (CPT1a) genes. Interestingly, the frequency of CPT1a + VDAC1 + M-MDSCs correlated positively with the severity of COVID-19 disease [65]. The significance of these findings is not clear. The upregulated expression of HKII and CPT1a genes suggests a distinct metabolic profile of MDSCs in COVID-19 characterized by activated glycolysis and fatty acid oxidation programs. VDAC1 is a mitochondrial membrane protein that functions as a major transporter of nucleotides and metabolites across the mitochondrial membrane [81]. Oligomerization of VDAC1 is involved in mitochondria-mediated apoptosis, leading to the release of mitochondrial DNA into the cytosol and subsequent induction of the type I IFN response [82,83]. Bone marrow-derived MDSCs and tumor-induced MDSCs have been shown to be able to produce type I IFN, which can participate in an autocrine loop of regulating PDL1 expression likely representing one of the ways to maintain their suppressive potential [83].
In conclusion, a considerable number of studies involving COVID-19 patients have reported the emergence of an immunosuppressive myeloid cell population with surface and molecular profiles similar to MDSCs in cancer patients. The number of these cells is increased in the lungs and bloodstream of COVID-19 patients, suggesting that they are involved in the regulating the local but also the systemic inflammatory immune response to SARS-CoV-2 infection.
4.2. MDSCs as correlates of the SARS-CoV-2 infection
Several studies have demonstrated that MDSC accumulation correlates with COVID-19 severity. For example, Sacchi et al. showed a higher percentage of PMN-MDSCs in patients who required intensive care unit (ICU) treatment compared to patients who did not require ICU treatment [71]. The same study also showed that high PMN-MDSC frequency at the admission could discriminate between survivors and non-survivors, further implicating MDSCs as indicators of COVID-19 severity [71]. A higher percentage of PMN-MDSCs and M-MDSCs was found in patients with ARDS as compared to patients with moderate disease [75]. Similarly, COVID-19 patients who developed ARDS showed a significant increase in PMN-MDSC values during hospitalization, whereas PMN-MDSCs did not rise in patients without ARDS [66]. Frequencies of PMN-MDSCs were significantly associated with disease severity in a study of 26 mild and 15 severe patients [70]. Falck-Jones et al. demonstrated that an early increase in M-MDSC frequency was associated with later disease severity [63]. A study by Tang et al., which enrolled 27 mild, 47 severe and 46 deceased COVID-19 patients, demonstrated that an M-MDSC cutoff value of ≥10%, as estimated in the CD14+ gate, predicted COVID-19 mortality [3]. On the other hand, a study by Mortaz et al. of 37 severe and 13 moderate COVID-19 patients showed that neither the frequency of M-MDSC nor PMN-MDSCs in peripheral blood predicted either the survival of patients or severity of infection [84]. In this study, no difference in PMN-MDSC frequency was found between ventilated and nonventilated patients, which is in contrast to the findings of Sacchi et al. [71,84]. The authors suggested that the observed discrepancies could be explained by differences in treatment and/or ethnicity between subjects of these two studies or possibly by the number of subjects analyzed [84]. Another possible explanation is differences in experimental design. In the study by Mortaz et al., PMN-MDSCs were analyzed in whole blood samples without density gradient centrifugation, which could lead to contamination with granulocytes, resulting in the identification of both MDSCs and granulocytes.
The literature is inconclusive regarding the correlation between the number of MDSCs and immunological parameters in COVID-19 patients. Several studies reported negative correlations between the frequencies of MDSCs and the absolute counts of T cells in COVID-19 patients [68,75,85,86]. There was also a negative correlation between the number of M-MDSCs and activated CD8+ T cells [73]. In another study of 147 COVID-19 patients, no correlation was found between blood MDSC frequencies and T cell counts in patients with COVID-19 either at the highest or lowest frequencies or at any of the longitudinal time points examined in each patient [63].
Some studies reported a negative correlation between MDSC frequencies and NK cells in COVID-19 patients, suggesting that MDSCs may be involved in the inhibition of this innate immune cell population in COVID-19 [85,86]. Two studies investigated correlations between MDSCs and inflammatory markers in patients with COVID-19. Emsen et al. reported a positive correlation between MDSCs and C-reactive protein, ferritin, and lactate dehydrogenase levels [70]. Xue et al. found that the MDSCs frequency was positively correlated with C-reactive protein and D-dimer [86].
In summary, almost all reported studies agree that MDSCs can be indicators of COVID-19 severity and the degree of systemic inflammation and lymphocyte exhaustion in patients with SARS-CoV-2 infection.
4.3. Factors involved in MDSC expansion and recruitment in COVID-19
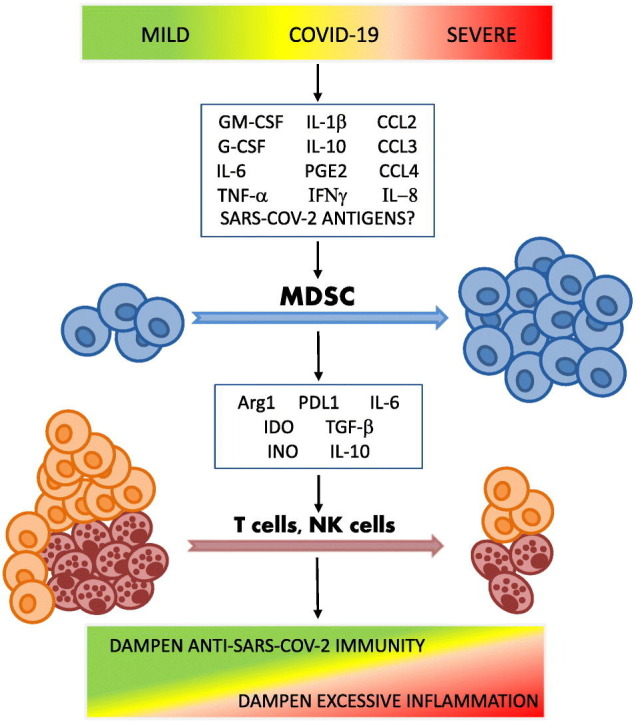
According to the two-stage model of MDSC development and expansion proposed by Gabrilovich et al, myelopoiesis of MDSCs in the bone marrow and their influx into the blood are driven by a number of inflammatory stimuli, such as IL-6, G-CSF, GM-CSF, vascular endothelial growth factor (VEGF) and possibly TNFα via activation of STAT3 and STAT5 signaling pathways. The establishment of the immunosuppressive profile of myeloid cells is then induced by factors such as IL-1β, IL-10, IFNγ, PGE2 and TLR ligands that activate STAT1, STAT6 and NF-κB transcription factors and upregulate cyclooxygenase 2 (COX2) [87]. Karin argues that there is a third type of signals required for successful mobilization of myeloid cells from the bone marrow or secondary lymphoid organs into the blood [74]. He proposed that this mobilization under cancer settings is orchestrated by chemokine receptors: C—C motif chemokine receptor 2 (CCR2) for M-MDSCs via the key ligand CCL2 and CCR5 for PMN-MDSCs via the key ligands: CCL3, CCL4 and CCL5. The simultaneous presence of all three types of signals necessary for MDSC development and expansion has been reported in COVID-19 patients.
Reizine et al. found an increase in plasma levels of IL-6, IL-10, G-CSF, and CCL2 along with an increase in MDSCs in COVID-19 patients [75]. The frequency of PMN-MDSCs in the peripheral blood correlated positively with IL-1β, IL-6, IL-8, and TNFα plasma levels [71]. A study by Takano et al. also reported a significant increase in IL-8 in severe COVID-19 patients. In this study, plasma levels of IL-8 correlated positively with the frequency of PMN-MDSCs, suggesting that IL-8 may be a specific chemokine signal responsible for the recruitment of MDSCs into peripheral circulation in COVID-19 [72]. A significant increase in the chemokines CCL2, CCL3, CCL4, which recruit myeloid cells, along with IL-6, IL-10, IFNγ, and TNFα was detected in the peripheral blood of severe COVID-19 patients compared to healthy controls [88]. Single-cell RNA sequencing revealed high expression of chemokines recruiting myeloid cells (CCL2, CCL3, CCL4, CCL7, CCL8, CXCL1, CXCL2, CXCL3 and especially IL-8) and low expression of chemokines recruiting T cells (CXCL9 and CXCL16) in the bronchoalveolar lavage of severe COVID-19, indicating preferential recruitment of myeloid cells to the lungs in COVID-19 patients [79,89].
High plasma levels of IL-6 and IL-10 in parallel with increased frequencies of M-MDSCs were reported in a study of 147 COVID-19 patients [63]. Consistent with this finding, cultivation of hematopoietic stem and progenitor cells isolated from human bone marrow with plasma from patients with severe COVID-19 stimulated generation of immunosuppressive CD14+ cells with increased expression of MS1 genes and decreased expression of MHC-II genes via IL-6 and IL-10, suggesting a key role of these cytokines in the formation of MDSCs in COVID-19 [90].
Strong correlations between disease severity and levels of the main MDSC-promoting mediators IL-6 and GM-CSF were demonstrated in a study of 471 COVID-19 patients [91]. Interestingly enough, elevated levels of GM-CSF appeared to be a distinctive feature of severe COVID-19, distinguishing it from fatal influenza [91] and likely uniquely contributing to the drastic changes in the myeloid cell compartment observed in COVID-19. Elevated serum levels of GM-CSF along with an increased incidence of MDCSs have also been reported in COVID–19 convalescents [68].
Not only host-derived mediators are involved in the development and expansion of MDSCs, but also some viral and bacterial antigens have been shown to trigger the generation of MDSCs. For example, hepatitis C virus core protein and hepatitis B virus surface antigen could induce generation of MDSCs from peripheral blood mononuclear cells via the phosphoinositide 3-kinase and ERK/IL-6/STAT3 pathways, respectively [92]. Xue et al. demonstrated a positive correlation between MDSC levels and SARS-CoV-2 viral load [86], suggesting that SARS-CoV-2 antigens may be involved in MDSC expansion. However, we have not found further evidence to support this assumption, so this remains a topic of discussion.
4.4. Functional characteristics of MDSCs in COVID-19
Lymphocytopenia, which affects mostly T cells and to a lesser extent NK and NKT cells but not B cells, is the hallmark of COVID-19 [76]. As described above, MDSCs utilize a number of immunosuppressive mechanisms, with T cells being their main target. Decreased proliferative capacity of CD8+ and CD4+ T cells and increased levels of apoptotic T cells have been noted in COVID-19 patients [65,75], likely contributing to the observed lymphocytopenia [63,93]. Several MDSC-mediated factors may cause T cell exhaustion in COVID-19. Arg1 production is one effector mechanism by which MDSCs suppress T cell proliferation via the degradation of L-arginine, which is critical for T cell proliferation and IFNγ production. Increased levels of Arg1 accompanied by significant decreases in L-arginine levels have been consistently observed in the plasma of COVID-19 patients, especially in severe cases [63,64,75,76,94]. Addition of L-arginine to MDSC co-cultures obtained from COVID-19 patients restored IFNγ production and provided partial restoration of T cell proliferation in vitro [63]. Moreover, plasmatic L-arginine in COVID-19 patients correlated negatively with the frequency of PMN-MDSCs, suggesting that MDSCs contribute to L-arginine deprivation in COVID-19 [94].
Another suppressive factor that may contribute to the decreased numbers of T cells in COVID-19 is IDO. IDO is able to induce activation of the kinase GCN2, leading to cell-cycle arrest and subsequent inhibition of T cell proliferation [95]. In COVID-19 patients, enhanced IDO activity correlated with lymphocytopenia, especially in ARDS patients [75]. The production of IDO by MDSCs is mainly, but not uniquely regulated by PGE2, the level of which is markedly elevated in the blood of COVID-19 patients [96]. Exposure to PGE2 has been shown to induce expression of endogenous COX2 in differentiating monocytes, leading to the establishment of a PGE2-COX2-mediated positive feedback loop and IDO induction [97]. Accordingly, a study by Tomić et al. showed increased expression of IDO by M-MDSCs from severe COVID-19 patients compared to healthy donors or mild COVID-19 patients [76].
In addition, PMN-MDSC from COVID-19 patients inhibited IFNγ production by T cells upon stimulation with SARS-CoV-2 peptides via TGFβ and NO, as anti-TGFβ and anti-iNOS treatments restored the cytokine-producing capacity of T cells [71]. The study also demonstrated a direct correlation between the PMN-MDSC frequencies and TGFβ in the plasma of COVID-19 patients, suggesting that PMN-MDSCs may contribute to TGFβ production. There is also evidence that increased expression of PDL1 and IL-10 in expanded MDSCs may contribute to T cell exhaustion in severe COVID-19 [76].
5. The role of MDSCs in COVID-19
A recent proteomic study suggested that the unique reshaping of the myeloid cell compartment observed in COVID-19 can be attributed to the SARS-CoV-2 non-structural protein 10. This viral protein interacts with the NF-κB repressing factor and triggers massive production of IL-6 and IL-8 by host cells, which simulate chemotaxis of myeloid cells [98]. Remodeling of the myeloid cell compartment in COVID-19 is marked not only by dramatic expansion of myeloid cells in the systemic circulation and airways of patients, but also by alterations in myeloid cell composition characterized by pronounced expansion of CD16+CD14+ nonclassical pro-inflammatory monocytes, dysfunctional mature neutrophils and MDSCs [5,99]. Infiltration with inflammatory monocytes and neutrophils is at least partially responsible for lung tissue damage in SARS-CoV-2 infection [89,100], whereas the role of MDSCs is not as clear.
Similar to cancer, MDSCs in COVID-19 patients are shaped by the coordinated action of mediators required for the initiation of emergency myelopoiesis and the activation and expansion of MDSCs. Positive correlations between the frequency of MDSCs and plasma levels of pro-inflammatory cytokines and other inflammatory markers suggest that MDSC expansion represents a systemic compensatory response to SARS-CoV-2 infection aimed at reducing excessive inflammation. On the other hand, assessment of the suppressive activity of MDSCs generated in response to SARS-CoV-2 infection showed that they are able to efficiently inhibit effector T cells via the production of Arg1, IDO, TGFβ and iNOS, suggesting that MDSCs have the potential to interfere with the elimination of the virus by T cells. Moreover, along with monocytes, M-MDSC and PMN-MDSC were shown to be the major producers of IL-6 in severe COVID-19 patients suggesting that they can potentially contribute to the exaggerated pro-inflammatory response and cytokine storm in COVID-19 patients [76]. The answer to the question of whether MDSCs are protective or destructive in the context of SARS-CoV-2 infection is complex and likely depends on the phase and severity of infection.
Some valuable insights into the role of MDSCs in infection-related systemic inflammation can be derived from adoptive transfer studies of sepsis. The elegant studies by Brudecki et al. and Derive et al. demonstrated a heterogeneous role of MDSCs in sepsis due to a phenotypic shift during infection [101,102]. Brudecki et al. showed that adoptive transfer of early-sepsis MDSCs (day 3 after the onset of infection) into mice with induced infection increased levels of pro-inflammatory cytokines (TNFα, IL-6) and worsened mortality, possibly via a cytokine storm and multi-organ failure [101]. In contrast, adoptive transfer of late-sepsis MDSCs (day 12) resulted in elevated serum levels of anti-inflammatory cytokines (IL-10, TGFβ) and reduced mortality [101]. Similarly, Derive et al. showed that adoptive transfer of MDSCs harvested on day 10 after the onset of sepsis dramatically improved the survival rate of infected mice, whereas MDSCs harvested on day 3 offered no protection [102]. In both studies, MDSCs were found to exhibit different phenotypes in terms of immunosuppressive factor production, responsiveness to stimuli, and the level of maturation depending on the stage of disease. However, both early- and late-stage MDSCs were able to effectively inhibit T cell proliferation [102]. These results suggest that the immunomodulatory activities of MDSCs are governed by the ongoing inflammatory process, and while early MDSCs are pro-inflammatory, late MDSCs may exert tolerogenic effects and contribute to the reduction of inflammation.
Taken together, these data may explain the conflicting results of several studies that have estimated the prognostic significance of MDSCs in COVID-19. For example, a study by Sacchi et al. showed that high MDSC frequencies at the admission could discriminate between survivors and non-survivors [71]. Whereas a study by Takano et al. showed that severe COVID-19 patients benefited from a transient increase in MDSCs in later stages of the disease. This study followed 13 patients with severe COVID-19 from symptom onset to discharge/death. They observed a delayed increase in PMN-MDSC frequencies in 8 recovered severe patients, while PMN-MDSCs remained unchanged in 5 non-survivors throughout the course of infection [72]. Another study by Jiménez-Cortegana et al. followed 40 severe discharged and 40 deceased patients. The results of the follow-up demonstrated steadily elevated counts of MDSCs until death in the peripheral blood of patients who eventually died. Discharged patients experienced a transitory increase in the counts of MDSCs during the first two weeks after ICU admission with a subsequent decline to baseline levels at discharge [103]. Similarly, Trombetta et a. showed that recovery of severe COVID-19 patients was associated with timely acquisition of a myeloid cell immunoregulatory phenotype [104]. It appears, that in the early phase of COVID-19, which is characterized by high viral loads in the system and high levels of numerous inflammatory mediators, progressive MDSC expansion inhibits protective immunity by suppressing SARS-CoV-2-specific T cells and probably NK cells, and possibly contributes to self-amplification of the inflammatory process through the production of pro-inflammatory cytokines such as IL-6. Whereas the spread of MDSCs in later stages of the disease may help restore a balance in the host, favoring resolution of inflammation and protecting organs from the damage by hyperactivated cytotoxic T cells. Similar to the sepsis studies, which have demonstrated that the functions and phenotype of MDSCs are highly dependent on the inflammatory environment, we can speculate that MDSCs arising at different stages of COVID-19 infection have different phenotypes and functions.
In conclusion, while the negative role of MDSCs in cancer is well documented and recognized, it is too early to draw the same conclusion about the role of MDSCs in COVID-19 patients, especially in patients with ARDS. Further studies using harmonized approaches to the identification of these cells are needed before the introduction of therapies targeting the activity of MDSCs.
Funding
This work was supported by the Committee of Science of the Ministry of Education and Science of the Republic of Kazakhstan [grant #AP09259390 “Study of the effect of SARS-CoV-2 antigens on the pro-inflammatory activity of neutrophils, monocytes and T regulatory cells”, 2021–2023 yy and grant #AP09259103 "Genomic and subgenomic characterization of SARS-CoV-2 strains circulating in Kazakhstan for the development of scientific approaches to COVID-19 diagnostics", 2021-2023 yy].
Declaration of interest statement
None of the authors has any potential financial conflict of interest related to this manuscript.
Acknowledgments
The authors thank Dr. Micceri, Ph.D for critical reading of the manuscript.
References
- 1.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rendeiro A.F., Casano J., Vorkas C.K., Singh H., Morales A., DeSimone R.A., Ellsworth G.B., Soave R., Kapadia S.N., Saito K., Brown C.D., Hsu J., Kyriakides C., Chui S., Cappelli L., Cacciapuoti M.T., Tam W., Galluzzi L., Simonson P.D., Elemento O., Salvatore M., Inghirami G. Longitudinal immune profiling of mild and severe COVID-19 reveals innate and adaptive immune dysfunction and provides an early prediction tool for clinical progression. MedRxiv. 2020 doi: 10.1101/2020.09.08.20189092. [DOI] [Google Scholar]
- 3.Tang G., Huang M., Luo Y., Liu W., Lin Q., Mao L., Wu S., Xiong Z., Hou H., Sun Z., Wang F. The dynamic immunological parameter landscape in coronavirus disease 2019 patients with different outcomes. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.697622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silvin A., Chapuis N., Dunsmore G., Goubet A.-G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N., Rameau P., Catelain C., Alfaro A., Dussiau C., Friedrich C., Sourdeau E., Marin N., Szwebel T.-A., Cantin D., Mouthon L., Borderie D., Deloger M., Bredel D., Mouraud S., Drubay D., Andrieu M., Lhonneur A.-S., Saada V., Stoclin A., Willekens C., Pommeret F., Griscelli F., Ng L.G., Zhang Z., Bost P., Amit I., Barlesi F., Marabelle A., Pène F., Gachot B., André F., Zitvogel L., Ginhoux F., Fontenay M., Solary E. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., De Domenico E., Wendisch D., Grasshoff M., Kapellos T.S., Beckstette M., Pecht T., Saglam A., Dietrich O., Mei H.E., Schulz A.R., Conrad C., Kunkel D., Vafadarnejad E., Xu C.-J., Horne A., Herbert M., Drews A., Thibeault C., Pfeiffer M., Hippenstiel S., Hocke A., Müller-Redetzky H., Heim K.-M., Machleidt F., Uhrig A., Bosquillon de Jarcy L., Jürgens L., Stegemann M., Glösenkamp C.R., Volk H.-D., Goffinet C., Landthaler M., Wyler E., Georg P., Schneider M., Dang-Heine C., Neuwinger N., Kappert K., Tauber R., Corman V., Raabe J., Kaiser K.M., Vinh M.T., Rieke G., Meisel C., Ulas T., Becker M., Geffers R., Witzenrath M., Drosten C., Suttorp N., von Kalle C., Kurth F., Händler K., Schultze J.L., Aschenbrenner A.C., Li Y., Nattermann J., Sawitzki B., Saliba A.-E., Sander L.E., Angelov A., Bals R., Bartholomäus A., Becker A., Bezdan D., Bonifacio E., Bork P., Clavel T., Colome-Tatche M., Diefenbach A., Dilthey A., Fischer N., Förstner K., Frick J.-S., Gagneur J., Goesmann A., Hain T., Hummel M., Janssen S., Kalinowski J., Kallies R., Kehr B., Keller A., Kim-Hellmuth S., Klein C., Kohlbacher O., Korbel J.O., Kurth I., Landthaler M., Li Y., Ludwig K., Makarewicz O., Marz M., McHardy A., Mertes C., Nöthen M., Nürnberg P., Ohler U., Ossowski S., Overmann J., Peter S., Pfeffer K., Poetsch A.R., Pühler A., Rajewsky N., Ralser M., Rieß O., Ripke S., Nunes da Rocha U., Rosenstiel P., Saliba A.-E., Sander L.E., Sawitzki B., Schiffer P., Schulte E.-C., Schultze J.L., Sczyrba A., Stegle O., Stoye J., Theis F., Vehreschild J., Vogel J., von Kleist M., Walker A., Walter J., Wieczorek D., Ziebuhr J. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veglia F., Sanseviero E., Gabrilovich D.I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 2021 doi: 10.1038/s41577-020-00490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koushki K., Salemi M., Miri S.M., Arjeini Y., Keshavarz M., Ghaemi A. Role of myeloid-derived suppressor cells in viral respiratory infections; hints for discovering therapeutic targets for COVID-19. Biomed. Pharmacother. 2021 doi: 10.1016/j.biopha.2021.112346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Santo C., Salio M., Masri S.H., Lee L.Y., Dong T., Speak A.O., Porubsky S., Booth S., Veerapen N., Besra G.S., Gröne H.J., Platt F.M., Zambon M., Cerundolo V. Invariant NKT cells reduce the immunosuppressive activity of influenza A virus-induced myeloid-derived suppressor cells in mice and humans. J. Clin. Invest. 2008;2008 doi: 10.1172/JCI36264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeisy-Scott V., Davis W.G., Patel J.R., Bowzard J.B., Shieh W.J., Zaki S.R., Katz J.M., Sambhara S. Increased MDSC accumulation and Th2 biased response to influenza A virus infection in the absence of TLR7 in mice. PLoS One. 2011 doi: 10.1371/journal.pone.0025242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fullerton J.N., O’Brien A.J., Gilroy D.W. Pathways mediating resolution of inflammation: when enough is too much. J. Pathol. 2013;231:8–20. doi: 10.1002/path.4232. [DOI] [PubMed] [Google Scholar]
- 11.COVID-19 mortality overview Stat. Nat. Cent. Heal. 2022. https://www.cdc.gov/nchs/covid19/mortality-overview.htm (n.d.) Accessed on February 23.
- 12.Simmonds P. Pervasive RNA secondary structure in the genomes of SARS-CoV-2 and other coronaviruses. MBio. 2020;11 doi: 10.1128/mBio.01661-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Li Y., Liu Q., Yao Q., Wang X., Zhang H., Chen R., Ren L., Min J., Deng F., Yan B., Liu L., Hu Z., Wang M., Zhou Y. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021;7:17. doi: 10.1038/s41421-021-00249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen I.-Y., Moriyama M., Chang M.-F., Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada T., Sato S., Sotoyama Y., Orba Y., Sawa H., Yamauchi H., Sasaki M., Takaoka A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021;22:820–828. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- 18.Yin X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., De Jesus P.D., Yang C.-C., Herbert K.M., Yoh S., Hultquist J.F., García-Sastre A., Chanda S.K. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramasamy S., Subbian S. Critical determinants of cytokine storm and type I interferon response in COVID-19 pathogenesis. Clin. Microbiol. Rev. 2021;34 doi: 10.1128/CMR.00299-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorne L.G., Reuschl A., Zuliani-Alvarez L., Whelan M.V.X., Turner J., Noursadeghi M., Jolly C., Towers G.J. SARS-CoV-2 sensing by RIGI and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 2021 doi: 10.15252/embj.2021107826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aboudounya M.M., Heads R.J. COVID-19 and toll-like receptor 4 (TLR4): SARS-CoV-2 may bind and activate TLR4 to increase ACE2 expression, facilitating entry and causing hyperinflammation. Mediat. Inflamm. 2021;2021:8874339. doi: 10.1155/2021/8874339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan P., Shen M., Yu Z., Ge W., Chen K., Tian M., Xiao F., Wang Z., Wang J., Jia Y., Wang W., Wan P., Zhang J., Chen W., Lei Z., Chen X., Luo Z., Zhang Q., Xu M., Li G., Li Y., Wu J. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun. 2021 doi: 10.1038/s41467-021-25015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L., Long X., Xu Q., Tan J., Wang G., Cao Y., Wei J., Luo H., Zhu H., Huang L., Meng F., Huang L., Wang N., Zhou X., Zhao L., Chen X., Mao Z., Chen C., Li Z., Sun Z., Zhao J., Wang D., Huang G., Wang W., Zhou J. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:992–994. doi: 10.1038/s41423-020-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scozzi D., Cano M., Ma L., Zhou D., Zhu J.H., O’Halloran J.A., Goss C., Rauseo A.M., Liu Z., Sahu S.K., Peritore V., Rocco M., Ricci A., Amodeo R., Aimati L., Ibrahim M., Hachem R., Kreisel D., Mudd P.A., Kulkarni H.S., Gelman A.E. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight. 2021;6 doi: 10.1172/jci.insight.143299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bime C., Casanova N.G., Nikolich-Zugich J., Knox K.S., Camp S.M., Garcia J.G.N. Strategies to DAMPen COVID-19-mediated lung and systemic inflammation and vascular injury. Transl. Res. 2021;232:37–48. doi: 10.1016/j.trsl.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015 doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Land W.G. Role of DAMPs in respiratory virus-induced acute respiratory distress syndrome—with a preliminary reference to SARS-CoV-2 pneumonia. Genes Immun. 2021 doi: 10.1038/s41435-021-00140-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coperchini F., Chiovato L., Rotondi M. Interleukin-6, CXCL10 and infiltrating macrophages in COVID-19-related cytokine storm: not one for all but all for one! Front. Immunol. 2021 doi: 10.3389/fimmu.2021.668507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou T., Su T.T., Mudianto T., Wang J. Immune asynchrony in COVID-19 pathogenesis and potential immunotherapies. J. Exp. Med. 2020;217 doi: 10.1084/jem.20200674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020 doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suthar M.S., Zimmerman M.G., Kauffman R.C., Mantus G., Linderman S.L., Hudson W.H., Vanderheiden A., Nyhoff L., Davis C.W., Adekunle O., Affer M., Sherman M., Reynolds S., Verkerke H.P., Alter D.N., Guarner J., Bryksin J., Horwath M.C., Arthur C.M., Saakadze N., Smith G.H., Edupuganti S., Scherer E.M., Hellmeister K., Cheng A., Morales J.A., Neish A.S., Stowell S.R., Frank F., Ortlund E., Anderson E.J., Menachery V.D., Rouphael N., Mehta A.K., Stephens D.S., Ahmed R., Roback J.D., Wrammert J. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep. Med. 2020 doi: 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stokes E.K., Zambrano L.D., Anderson K.N., Marder E.P., Raz K.M., El Burai Felix S., Tie Y., Fullerton K.E., Disease Coronavirus. Case surveillance - United States, January 22-May 30, 2020. MMWR Morb. Mortal. Wkly Rep. 2019;69(2020):759–765. doi: 10.15585/mmwr.mm6924e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gold M.S., Sehayek D., Gabrielli S., Zhang X., McCusker C., Ben-Shoshan M. COVID-19 and comorbidities: a systematic review and meta-analysis. Postgrad. Med. 2020 doi: 10.1080/00325481.2020.1786964. [DOI] [PubMed] [Google Scholar]
- 35.Latif M.B., Shukla S., Del Rio Estrada P.M., Ribeiro S.P., Sekaly R.P., Sharma A.A. Immune mechanisms in cancer patients that lead to poor outcomes of SARS-CoV-2 infection. Transl. Res. 2022;241:83–95. doi: 10.1016/j.trsl.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamyshnyi A., Krynytska I., Matskevych V., Marushchak M., Lushchak O. Arterial hypertension as a risk comorbidity associated with COVID-19 pathology. Int. J. Hypertens. 2020;2020:1–7. doi: 10.1155/2020/8019360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bajaj V., Gadi N., Spihlman A.P., Wu S.C., Choi C.H., Moulton V.R. Aging, immunity, and COVID-19: how age influences the host immune response to coronavirus infections? Front. Physiol. 2021;11 doi: 10.3389/fphys.2020.571416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., Pellegrini K.L., Manfredi C., Sorscher E., Mainou B., Lobby J.L., Kohlmeier J.E., Lowen A.C., Shi P.-Y., Menachery V.D., Anderson L.J., Grakoui A., Bosinger S.E., Suthar M.S. Type I and Type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. 2020;94 doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatton C.F., Botting R.A., Dueñas M.E., Haq I.J., Verdon B., Thompson B.J., Spegarova J.S., Gothe F., Stephenson E., Gardner A.I., Murphy S., Scott J., Garnett J.P., Carrie S., Powell J., Khan C.M.A., Huang L., Hussain R., Coxhead J., Davey T., Simpson A.J., Haniffa M., Hambleton S., Brodlie M., Ward C., Trost M., Reynolds G., Duncan C.J.A. Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2. Nat. Commun. 2021;12 doi: 10.1038/S41467-021-27318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Liang Q.-Z., Lu W., Yang Y.-L., Chen R., Huang Y.-W., Wang B. A comparative analysis of coronavirus nucleocapsid (N) proteins reveals the SADS-CoV N protein antagonizes IFN-β production by inducing ubiquitination of RIG-I. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.688758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017;91 doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spiegel M., Pichlmair A., Martínez-Sobrido L., Cros J., García-Sastre A., Haller O., Weber F. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong H.H., Fung T.S., Fang S., Huang M., Le M.T., Liu D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barth R.F., Buja L.M., Parwani A.V. The spectrum of pathological findings in coronavirus disease (COVID-19) and the pathogenesis of SARS-CoV-2. Diagn. Pathol. 2020;15:85. doi: 10.1186/s13000-020-00999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alipoor S.D., Jamaati H., Tabarsi P., Mortaz E. Immunopathogenesis of pneumonia in COVID-19. Tanaffos. 2020;19:79–82. [PMC free article] [PubMed] [Google Scholar]
- 47.Marais C., Claude C., Semaan N., Charbel R., Barreault S., Travert B., Piloquet J.-E., Demailly Z., Morin L., Merchaoui Z., Teboul J.-L., Durand P., Miatello J., Tissières P., Barreault S., Beggaz M., Charbel R., Claude C., Demailly Z., Durand P., Gerschenfeld G., Giraldi J., Guerra M., Hily M., Journaux M., Lai C., Leroux P., Marais C., Merchaoui Z., Miatello J., Niçaise C., Piloquet J.-E., Ren M., Simbozel M., Semaan N., Teboul J.-L., Tissieres P., Travert B. Myeloid phenotypes in severe COVID-19 predict secondary infection and mortality: a pilot study. Ann. Intensive Care. 2021;11:111. doi: 10.1186/s13613-021-00896-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veglia F., Perego M., Gabrilovich D. Myeloid-derived suppressor cells coming of age review-article. Nat. Immunol. 2018;19:108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostrand-Rosenberg S., Fenselau C. Myeloid-derived suppressor cells: immune-suppressive cells that impair antitumor immunity and are sculpted by their environment. J. Immunol. 2018;200:422–431. doi: 10.4049/jimmunol.1701019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bronte V., Brandau S., Chen S.H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S., Rodriguez P.C., Sica A., Umansky V., Vonderheide R.H., Gabrilovich D.I. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016;7:1–13. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan A.N.H., Emmons T.R., Wong J.T., Alqassim E., Singel K.L., Mark J., Smith B.E., Tario J.D., Eng K.H., Moysich K.B., Odunsi K., Abrams S.I., Segal B.H. Quantification of early-stage myeloid-derived suppressor cells in cancer requires excluding basophils. Cancer Immunol. Res. 2020;8:819–828. doi: 10.1158/2326-6066.CIR-19-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Condamine T., Dominguez G.A., Youn J.I., Kossenkov A.V., Mony S., Alicea-Torres K., Tcyganov E., Hashimoto A., Nefedova Y., Lin C., Partlova S., Garfall A., Vogl D.T., Xu X., Knight S.C., Malietzis G., Lee G.H., Eruslanov E., Albelda S.M., Wang X., Mehta J.L., Bewtra M., Rustgi A., Hockstein N., Witt R., Masters G., Nam B., Smirnov D., Sepulveda M.A., Gabrilovich D.I. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 2016;1:1–12. doi: 10.1126/sciimmunol.aaf8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alshetaiwi H., Pervolarakis N., McIntyre L.L., Ma D., Nguyen Q., Rath J.A., Nee K., Hernandez G., Evans K., Torosian L., Silva A., Walsh C., Kessenbrock K. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.aay6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cassetta L., Bruderek K., Skrzeczynska-Moncznik J., Osiecka O., Hu X., Rundgren I.M., Lin A., Santegoets K., Horzum U., Godinho-Santos A., Zelinskyy G., Garcia-Tellez T., Bjelica S., Taciak B., Kittang A.O., Höing B., Lang S., Dixon M., Müller V., Utikal J.S., Karakoç D., Yilmaz K.B., Górka E., Bodnar L., Anastasiou O.E., Bourgeois C., Badura R., Kapinska-Mrowiecka M., Gotic M., ter Laan M., Kers-Rebel E., Król M., Santibañez J.F., Müller-Trutwin M., Dittmer U., de Sousa A.E., Esendağlı G., Adema G., Loré K., Ersvær E., Umansky V., Pollard J.W., Cichy J., Brandau S. Differential expansion of circulating human MDSC subsets in patients with cancer, infection and inflammation. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2020-001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruno A., Mortara L., Baci D., Noonan D.M., Albini A. Myeloid derived suppressor cells interactions with natural killer cells and pro-angiogenic activities: roles in tumor progression. Front. Immunol. 2019;10:771. doi: 10.3389/fimmu.2019.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Özkan B., Lim H., Park S.-G. Immunomodulatory function of myeloid-derived suppressor cells during B cell-mediated immune responses. Int. J. Mol. Sci. 2018;19:1468. doi: 10.3390/ijms19051468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ge Y., Cheng D., Jia Q., Xiong H., Zhang J. Mechanisms underlying the role of myeloid-derived suppressor cells in clinical diseases: good or bad. Immune Netw. 2021;21 doi: 10.4110/in.2021.21.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohl K., Tenbrock K. Reactive oxygen species as regulators of MDSC-mediated immune suppression. Front. Immunol. 2018;9:2499. doi: 10.3389/fimmu.2018.02499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nagaraj S., Gupta K., Pisarev V., Kinarsky L., Sherman S., Kang L., Herber D.L., Schneck J., Gabrilovich D.I. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat. Med. 2007;13:828–835. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez P.C., Quiceno D.G., Ochoa A.C. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood. 2007;109:1568–1573. doi: 10.1182/blood-2006-06-031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pyzer A.R., Cole L., Rosenblatt J., Avigan D.E. Myeloid-derived suppressor cells as effectors of immune suppression in cancer. Int. J. Cancer. 2016;139:1915–1926. doi: 10.1002/ijc.30232. [DOI] [PubMed] [Google Scholar]
- 62.Noman M.Z., Desantis G., Janji B., Hasmim M., Karray S., Dessen P., Bronte V., Chouaib S. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced: MDSC-mediated T cell activation. J. Exp. Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Falck-Jones S., Vangeti S., Yu M., Falck-Jones R., Cagigi A., Badolati I., Österberg B., Lautenbach M.J., Åhlberg E., Lin A., Lepzien R., Szurgot I., Lenart K., Hellgren F., Maecker H., Sälde J., Albert J., Johansson N., Bell M., Loré K., Färnert A., Smed-Sörensen A. Functional monocytic myeloid-derived suppressor cells increase in blood but not airways and predict COVID-19 severity. J. Clin. Invest. 2021;131 doi: 10.1172/JCI144734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dean M.J., Ochoa J.B., Sanchez-Pino M., Zabaleta J., Garai J., Del Valle L., Wyczechowska D., Buckner L., Philbrook P., Majumder R., Vander Heide R., Dunkenberger L., Thylur R., Nossaman R., Roberts W.M., Chapple A., Collins J., Luke B., Johnson R., Koul H., Rees C.A., Morris C.R., Garcia-Diaz J., Ochoa A.C. Transcriptome and functions of granulocytic myeloid-derived suppressor cells determine their association with disease severity of COVID-19. MedRxiv Prepr. Serv. Heal. Sci. 2021 doi: 10.1101/2021.03.26.21254441. [DOI] [Google Scholar]
- 65.Thompson E.A., Cascino K., Ordonez A.A., Zhou W., Vaghasia A., Hamacher-Brady A., Brady N.R., Sun I.-H., Wang R., Rosenberg A.Z., Delannoy M., Rothman R., Fenstermacher K., Sauer L., Shaw-Saliba K., Bloch E.M., Redd A.D., Tobian A.A.R., Horton M., Smith K., Pekosz A., D’Alessio F.R., Yegnasubramanian S., Ji H., Cox A.L., Powell J.D. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coudereau R., Waeckel L., Cour M., Rimmele T., Pescarmona R., Fabri A., Jallades L., Yonis H., Gossez M., Lukaszewicz A.-C., Argaud L., Venet F., Monneret G. Emergence of immunosuppressive LOX-1+ PMN-MDSC in septic shock and severe COVID-19 patients with acute respiratory distress syndrome. J. Leukoc. Biol. 2022;111:489–496. doi: 10.1002/JLB.4COVBCR0321-129R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Agrati C., Sacchi A., Bordoni V., Cimini E., Notari S., Grassi G., Casetti R., Tartaglia E., Lalle E., D’Abramo A., Castilletti C., Marchioni L., Shi Y., Mariano A., Song J.-W., Zhang J.-Y., Wang F.-S., Zhang C., Fimia G.M., Capobianchi M.R., Piacentini M., Antinori A., Nicastri E., Maeurer M., Zumla A., Ippolito G. Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (COVID-19) Cell Death Differ. 2020;27:3196–3207. doi: 10.1038/s41418-020-0572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siemińska I., Węglarczyk K., Surmiak M., Kurowska-Baran D., Sanak M., Siedlar M., Baran J. Mild and asymptomatic COVID-19 convalescents present long-term endotype of immunosuppression associated with neutrophil subsets possessing regulatory functions. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.748097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ryan F.J., Hope C.M., Masavuli M.G., Lynn M.A., Mekonnen Z.A., Yeow A.E.L., Garcia-Valtanen P., Al-Delfi Z., Gummow J., Ferguson C., O’Connor S., Reddi B.A.J., Hissaria P., Shaw D., Kok-Lim C., Gleadle J.M., Beard M.R., Barry S.C., Grubor-Bauk B., Lynn D.J. Long-term perturbation of the peripheral immune system months after SARS-CoV-2 infection. BMC Med. 2022;20 doi: 10.1186/S12916-021-02228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emsen A., Sumer S., Tulek B., Cizmecioglu H., Vatansev H., Goktepe M.H., Kanat F., Koksal Y., Arslan U., Artac H. Correlation of myeloid-derived suppressor cells with C-reactive protein, ferritin and lactate dehydrogenase levels in patients with severe COVID-19. Scand. J. Immunol. 2022;95 doi: 10.1111/sji.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sacchi A., Grassi G., Bordoni V., Lorenzini P., Cimini E., Casetti R., Tartaglia E., Marchioni L., Petrosillo N., Palmieri F., D’Offizi G., Notari S., Tempestilli M., Capobianchi M.R., Nicastri E., Maeurer M., Zumla A., Locatelli F., Antinori A., Ippolito G., Agrati C. Early expansion of myeloid-derived suppressor cells inhibits SARS-CoV-2 specific T-cell response and may predict fatal COVID-19 outcome. Cell Death Dis. 2020;11:921. doi: 10.1038/s41419-020-03125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takano T., Matsumura T., Adachi Y., Terahara K., Moriyama S., Onodera T., Nishiyama A., Kawana-Tachikawa A., Miki S., Hosoya-Nakayama K., Nakamura-Hoshi M., Seki S., Tachikawa N., Yoshimura Y., Miyata N., Horiuchi H., Sasaki H., Miyazaki K., Kinoshita N., Sudo T., Akiyama Y., Sato R., Suzuki T., Matano T., Takahashi Y. Myeloid cell dynamics correlating with clinical outcomes of severe COVID-19 in Japan. Int. Immunol. 2021 doi: 10.1093/intimm/dxab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiménez-Cortegana C., Liró J., Palazón-Carrión N., Salamanca E., Sojo-Dorado J., de la Cruz-Merino L., Pascual Á., Rodríguez-Baño J., Sánchez-Margalet V. Increased blood monocytic myeloid derived suppressor cells but low regulatory T lymphocytes in patients with mild COVID-19. Viral Immunol. 2021;34:639–645. doi: 10.1089/vim.2021.0044. [DOI] [PubMed] [Google Scholar]
- 74.Karin N. The development and homing of myeloid-derived suppressor cells: from a two-stage model to a multistep narrative. Front. Immunol. 2020 doi: 10.3389/fimmu.2020.557586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reizine F., Lesouhaitier M., Gregoire M., Pinceaux K., Gacouin A., Maamar A., Painvin B., Camus C., Le Tulzo Y., Tattevin P., Revest M., Le Bot A., Ballerie A., Cador-Rousseau B., Lederlin M., Lebouvier T., Launey Y., Latour M., Verdy C., Rossille D., Le Gallou S., Dulong J., Moreau C., Bendavid C., Roussel M., Cogne M., Tarte K., Tadié J.-M. SARS-CoV-2-induced ARDS associates with MDSC expansion, lymphocyte dysfunction, and arginine shortage. J. Clin. Immunol. 2021;41:515–525. doi: 10.1007/s10875-020-00920-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomić S., Đokić J., Stevanović D., Ilić N., Gruden-Movsesijan A., Dinić M., Radojević D., Bekić M., Mitrović N., Tomašević R., Mikić D., Stojanović D., Čolić M. Reduced expression of autophagy markers and expansion of myeloid-derived suppressor cells correlate with poor T cell response in severe COVID-19 patients. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.614599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tsang O. Tak-Yin, Wagh D., Coller J., Pellegrini K.L., Kazmin D., Alaaeddine G., Leung W.S., Chan J.M.C., Chik T.S.H., Choi C.Y.C., Huerta C., McCullough M. Paine, Lv H., Anderson E., Edupuganti S., Upadhyay A.A., Bosinger S.E., Maecker H.T., Khatri P., Rouphael N., Peiris M., Pulendran B. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu G., Qi F., Li H., Yang Q., Wang H., Wang X., Liu X., Zhao J., Liao X., Liu Y., Liu L., Zhang S., Zhang Z. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020;6:73. doi: 10.1038/s41421-020-00225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shoshan-Barmatz V., De Pinto V., Zweckstetter M., Raviv Z., Keinan N., Arbel N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol. Asp. Med. 2010;31:227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 81.Ben-Hail D., Begas-Shvartz R., Shalev M., Shteinfer-Kuzmine A., Gruzman A., Reina S., De Pinto V., Shoshan-Barmatz V. Novel compounds targeting the mitochondrial protein VDAC1 inhibit apoptosis and protect against mitochondrial dysfunction. J. Biol. Chem. 2016;291:24986–25003. doi: 10.1074/jbc.M116.744284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J., Gupta R., Blanco L.P., Yang S., Shteinfer-Kuzmine A., Wang K., Zhu J., Yoon H.E., Wang X., Kerkhofs M., Kang H., Brown A.L., Park S.J., Xu X., van Rilland E.Z., Kim M.K., Cohen J.I., Kaplan M.J., Shoshan-Barmatz V., Chung J.H. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science. 2019 doi: 10.1126/science.aav4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiao W., Klement J.D., Lu C., Ibrahim M.L., Liu K. IFNAR1 controls autocrine type I IFN regulation of PD-L1 expression in myeloid-derived suppressor cells. J. Immunol. 2018 doi: 10.4049/jimmunol.1800129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mortaz E., Dezfuli N.K., Roofchayee N.D., Adcock I. MedRxiv. Cold Spring Harbor Laboratory Press; 2021. Myeloid-derived suppressor cells in the blood of COVID-19 patients; p. PA2318. [DOI] [Google Scholar]
- 85.Bordoni V., Sacchi A., Cimini E., Notari S., Grassi G., Tartaglia E., Casetti R., Giancola M.L., Bevilacqua N., Maeurer M., Zumla A., Locatelli F., De Benedetti F., Palmieri F., Marchioni L., Capobianchi M.R., D’Offizi G., Petrosillo N., Antinori A., Nicastri E., Ippolito G., Agrati C. An inflammatory profile correlates with decreased frequency of cytotoxic cells in coronavirus disease 2019. Clin. Infect. Dis. 2020;71:2272–2275. doi: 10.1093/cid/ciaa577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xue G., Jiang M., Zhao R., Le A., Li J. Elevated frequencies of CD14+HLA-DRlo/neg MDSCs in COVID-19 patients. Aging (Albany NY) 2021;13:6236–6246. doi: 10.18632/aging.202571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Condamine T., Gabrilovich D.I. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32:19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vanderbeke L., Van Mol P., Van Herck Y., De Smet F., Humblet-Baron S., Martinod K., Antoranz A., Arijs I., Boeckx B., Bosisio F.M., Casaer M., Dauwe D., De Wever W., Dooms C., Dreesen E., Emmaneel A., Filtjens J., Gouwy M., Gunst J., Hermans G., Jansen S., Lagrou K., Liston A., Lorent N., Meersseman P., Mercier T., Neyts J., Odent J., Panovska D., Penttila P.A., Pollet E., Proost P., Qian J., Quintelier K., Raes J., Rex S., Saeys Y., Sprooten J., Tejpar S., Testelmans D., Thevissen K., Van Buyten T., Vandenhaute J., Van Gassen S., Velásquez Pereira L.C., Vos R., Weynand B., Wilmer A., Yserbyt J., Garg A.D., Matthys P., Wouters C., Lambrechts D., Wauters E., Wauters J. Monocyte-driven atypical cytokine storm and aberrant neutrophil activation as key mediators of COVID-19 disease severity. Nat. Commun. 2021;12:4117. doi: 10.1038/s41467-021-24360-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Park J.H., Lee H.K. Re-analysis of single cell transcriptome reveals that the NR3C1-CXCL8-neutrophil axis determines the severity of COVID-19. Front. Immunol. 2020;11:2145. doi: 10.3389/fimmu.2020.02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reyes M., Filbin M.R., Bhattacharyya R.P., Sonny A., Mehta A., Billman K., Kays K.R., Pinilla-Vera M., Benson M.E., Cosimi L.A., Hung D.T., Levy B.D., Villani A.-C., Sade-Feldman M., Baron R.M., Goldberg M.B., Blainey P.C., Hacohen N. Plasma from patients with bacterial sepsis or severe COVID-19 induces suppressive myeloid cell production from hematopoietic progenitors in vitro. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abe9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thwaites R.S., Uruchurtu A. Sanchez Sevilla, Siggins M.K., Liew F., Russell C.D., Moore S.C., Fairfield C., Carter E., Abrams S., Short C.-E., Thaventhiran T., Bergstrom E., Gardener Z., Ascough S., Chiu C., Docherty A.B., Hunt D., Crow Y.J., Solomon T., Taylor G.P., Turtle L., Harrison E.M., Dunning J., Semple M.G., Baillie J.K., Openshaw P.J.M., ISARIC4C investigators Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abg9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O’Connor M.A., Rastad J.L., Green W.R. The role of myeloid-derived suppressor cells in viral infection. Viral Immunol. 2017;30:82–97. doi: 10.1089/vim.2016.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liao D., Zhou F., Luo L., Xu M., Wang H., Xia J., Gao Y., Cai L., Wang Z., Yin P., Wang Y., Tang L., Deng J., Mei H., Hu Y. Haematological characteristics and risk factors in the classification and prognosis evaluation of COVID-19: a retrospective cohort study. Lancet Haematol. 2020 doi: 10.1016/S2352-3026(20)30217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sacchi A., Grassi G., Notari S., Gili S., Bordoni V., Tartaglia E., Casetti R., Cimini E., Mariotti D., Garotto G., Beccacece A., Marchioni L., Bibas M., Nicastri E., Ippolito G., Agrati C. Expansion of myeloid derived suppressor cells contributes to platelet activation by L-arginine deprivation during SARS-CoV-2 infection. Cells. 2021;10:2111. doi: 10.3390/cells10082111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Munn D.H., Sharma M.D., Baban B., Harding H.P., Zhang Y., Ron D., Mellor A.L. GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3-dioxygenase. Immunity. 2005;22:633–642. doi: 10.1016/j.immuni.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 96.Ricke-Hoch M., Stelling E., Lasswitz L., Gunesch A.P., Kasten M., Zapatero-Belinchón F.J., Brogden G., Gerold G., Pietschmann T., Montiel V., Balligand J.-L., Facciotti F., Hirsch E., Gausepohl T., Elbahesh H., Rimmelzwaan G.F., Höfer A., Kühnel M.P., Jonigk D., Eigendorf J., Tegtbur U., Mink L., Scherr M., Illig T., Schambach A., Pfeffer T.J., Hilfiker A., Haverich A., Hilfiker-Kleiner D. Impaired immune response mediated by prostaglandin E2 promotes severe COVID-19 disease. PLoS One. 2021;16 doi: 10.1371/journal.pone.0255335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Obermajer N., Kalinski P. Generation of myeloid-derived suppressor cells using prostaglandin E2. Transp. Res. 2012;1:15. doi: 10.1186/2047-1440-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., Wang C., Zhang Y., Zhu Z., Liu Z., Peng C., Zhu T., Liang Q. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. 2021;2:99–112.e7. doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Müller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U.G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L.-E., Eils R. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 100.Miyazawa M. Immunopathogenesis of SARS-CoV-2-induced pneumonia: lessons from influenza virus infection. Inflamm. Regen. 2020;40:39. doi: 10.1186/s41232-020-00148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brudecki L., Ferguson D.A., McCall C.E., El Gazzar M. Myeloid-derived suppressor cells evolve during sepsis and can enhance or attenuate the systemic inflammatory response. Infect. Immun. 2012;80:2026–2034. doi: 10.1128/IAI.00239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Derive M., Bouazza Y., Alauzet C., Gibot S. Myeloid-derived suppressor cells control microbial sepsis. Intensive Care Med. 2012;38:1040–1049. doi: 10.1007/s00134-012-2574-4. [DOI] [PubMed] [Google Scholar]
- 103.Jiménez-Cortegana C., Sánchez-Jiménez F., Pérez-Pérez A., Álvarez N., Sousa A., Cantón-Bulnes L., Vilariño-García T., Fuentes S., Martín S., Jiménez M., León-Justel A., de la Cruz-Merino L., Garnacho-Montero J., Sánchez-Margalet V. Low levels of granulocytic myeloid-derived suppressor cells may be a good marker of survival in the follow-up of patients with severe COVID-19. Front. Immunol. 2022;12 doi: 10.3389/fimmu.2021.801410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Trombetta A.C., Farias G.B., Gomes A.M.C., Godinho-Santos A., Rosmaninho P., Conceição C.M., Laia J., Santos D.F., Almeida A.R.M., Mota C., Gomes A., Serrano M., Veldhoen M., Sousa A.E., Fernandes S.M. Severe COVID-19 recovery is associated with timely acquisition of a myeloid cell immune-regulatory phenotype. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.691725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Atanackovic D., Avila S.V., Lutfi F., de Miguel-Perez D., Fan X., Sanchez-Petitto G., Vander Mause E., Siglin J., Baddley J., Mannuel H.D., Alkhaldi H., Hankey K.G., Lapidus R., Kleinberg M., Rabin J., Shanholtz C., Rolfo C., Rapoport A.P., Dahiya S., Luetkens T. Deep dissection of the antiviral immune profile of patients with COVID-19. Commun. Biol. 2021;4 doi: 10.1038/s42003-021-02852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


