Figure S4.
The liquid phase is essential for incorporation of oskar mRNA in vitro, related to Figure 3
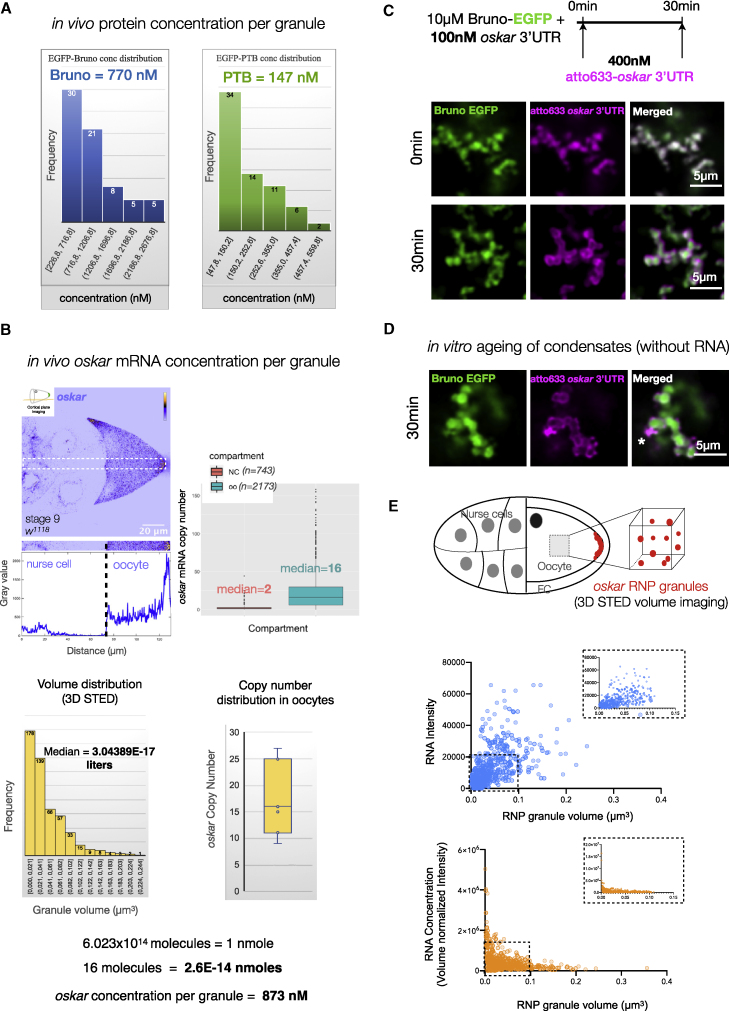
(A) Quantification of in vivo protein concentrations per granule. GFP-trap lines of Bruno and PTB were used and absolute concentrations calculated based on a calibration curve of recombinant EGFP imaged under identical conditions in the same imaging session (Xing et al., 2020). Numbers in the histogram refer to the mean number of granules grouped under the indicated range of concentration.
(B) For in vivo oskar RNA concentration per granule, w1118 egg chambers were stained for oskar by smFISH, and oskar copy number per granule in the oocyte compartment was calculated. A representative oskar smFISH image of a cortical plane acquisition done in “photon-counting mode” to avoid saturation of the signal in the oocyte. The intensity profile of the boxed area indeed shows the increase in oskar signal intensity along the AP axis. Granule volume obtained from 3D STED experiments was plotted, and absolute molar concentration of oskar RNA per granule was then derived based on average granule volume. Numbers in the histogram refer to the number of granules grouped under the indicated range of volume.
(C) Representative light microscopy single plane confocal images of experimental conditions used for cryoelectron tomography in Figure 3B. Images were acquired and processed independently.
(D) Condensates with Bruno alone preclude incorporation of oskar 3′UTR. Condensates were assembled with Bruno alone in 150 mM NaCl assay buffer. 10 nM atto633 labeled oskar 3′UTR RNA was added at 30 min of condensate aging. Note that new condensates formed after RNA addition show colocalization of the RNA and protein (marked by ∗).
(E) Plot of mRNA intensity versus granule volume of oskar RNP granules measured by 3D STED on w1118 egg chambers probed for oskar mRNA by smFISH. Intensity of oskar mRNA signal (top plot) was normalized by granule volume to derive RNA concentration per granule, which does not increase with increase in granule volume.