Figure 1.
Antigenic and structural characterization of homotypic and chimeric RBD-dimers
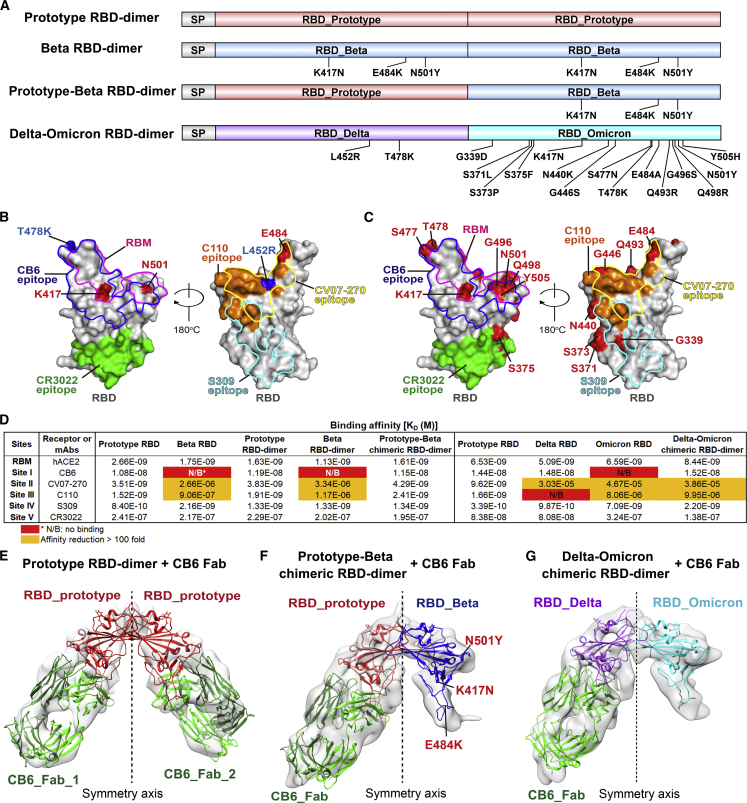
(A) Schematic diagram of prototype RBD-dimer, Beta RBD-dimer, prototype-Beta chimeric RBD-dimer, and Delta-Omicron chimeric RBD-dimer. Two SARS-CoV-2 RBDs were dimerized as tandem repeat (SP, signal peptide).
(B) Footprint of hACE2 and five classes of antibodies on SARS-CoV-2 RBD. Prototype RBD is shown as gray surface (PDB: 6LZG). Residues that mutated in SARS-CoV-2 Beta and Delta variants are colored in red and blue, respectively. Footprints of hACE2 (PDB: 6LZG), CB6 (PDB: 7C01), CV07-270 (PDB: 6XKP), C110 (PDB: 7K8V), S309 (PDB: 6WPT), and CR3022 (PDB: 6W41) are highlighted in pink, blue, yellow, orange, cyan, and green, respectively.
(C) Footprint of hACE2 and five classes of antibodies on SARS-CoV-2 RBD. Prototype RBD is shown as gray surface. Residues that mutated in SARS-CoV-2 Omicron variants are colored in red.
(D) Binding affinities of antigens bound to hACE2 and representative mAbs targeting five major sites. Red indicates no binding (N/B). The ones with affinity reductions more than 100-fold are colored in yellow.
(E) Density map of prototype RBD-dimer bound to two CB6 Fabs, with the atomic models of the SARS-CoV-2 RBD/CB6 complex (PDB:7C01) were fitted and rebuilt.
(F) Density map of prototype-Beta chimeric RBD-dimer bound to one CB6 Fab, with the atomic models of SARS-CoV-2 RBD (PDB: 6LZG) and RBD/CB6 complex (PDB: 7C01) were fitted and rebuilt, clearly showing that Beta variant RBD does not bind to CB6.
(G) Density map of Delta-Omicron chimeric RBD-dimer bound to one CB6 Fab, with the atomic models of Delta RBD (PDB: 7V8B), Omicron RBD (PDB: 7WBL) and CB6 (PDB: 7C01) were fitted and rebuilt, clearly showing that Omicron variant RBD does not bind to CB6.