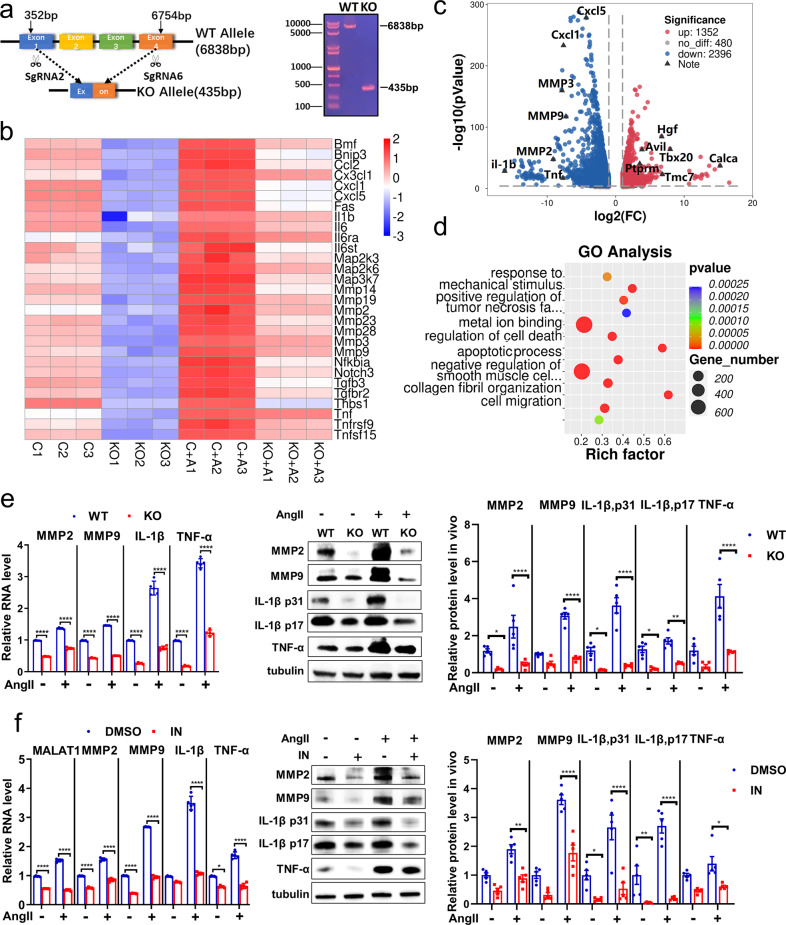
Fig. 6.
Inhibiting Malat1 can remarkably protect smooth muscle cells from AngII stimulation in vitro. a The monoclonal MOVAS cells with stable knockout of the Malat1 gene using the CRISPR/Cas9 system via lentiviral transfection were screened. b RNA transcriptome sequencing of Malat1−/− and WT MOVAS cells at baseline and with AngII stimulation. A heatmap of downregulated genes showed relative expression (z score) levels of abundantly and differentially expressed genes. c The fold change of all genes with a significant difference was presented in the volcano plot. d GO analysis of downregulated genes in Malat1−/− MOVAS cells compared with the WT type. e Qualitative PCR analysis of RNA levels of MMP2, MMP9, IL-1β, and TNF-α in KO and WT MOVAS cells at baseline and with AngII stimulation. n = 5 per group. Corresponding proteins were detected by immunoblotting. n = 5 per group. f The relative RNA levels of Malat1, MMPs, and inflammatory factors were detected in DMSO and MALAT1-IN-1 groups administered Ang II. n = 5 per group. Corresponding proteins were detected by immunoblotting. n = 5 per group. Data were presented as mean ± SEM and normal distributions were tested by the Shapiro–Wilk method, which showed that all data were normally distributed. Two-way ANOVA followed by Sidak post hoc test used for (e, f). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Each experiment was repeated independently for a minimum of three times