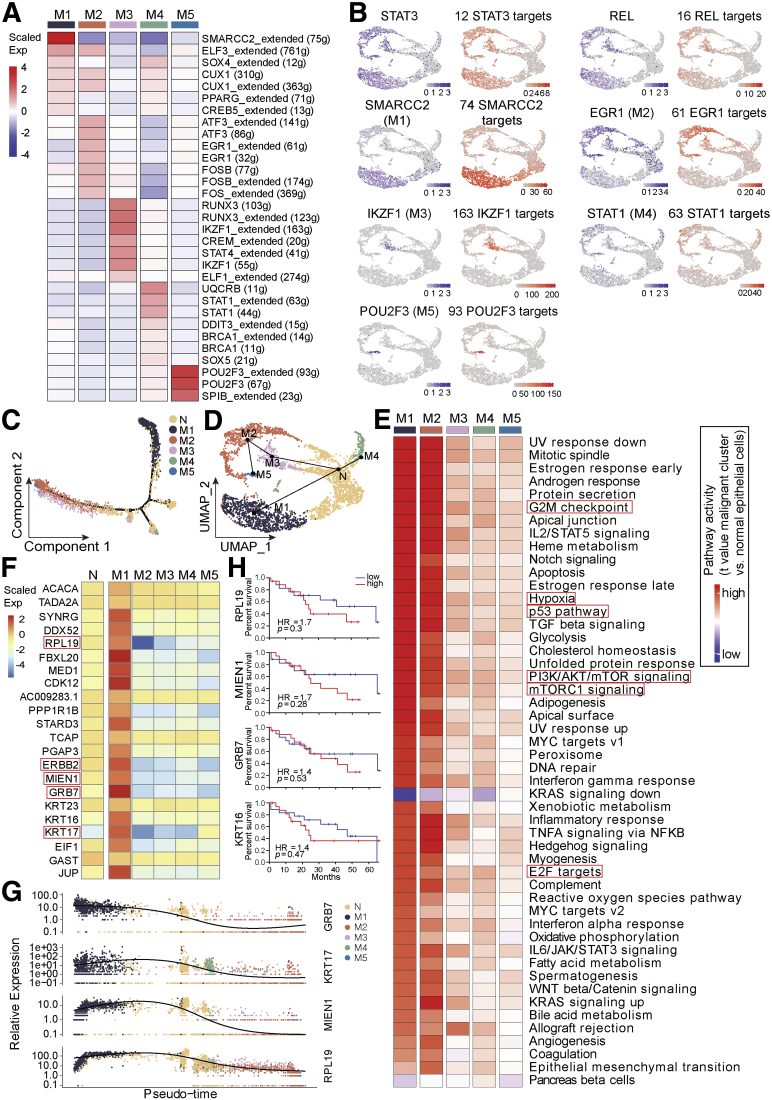
Figure 5.
Transcriptomic heterogeneity of malignant cells in human dCCA tissues.A, Heat map of the t-value for the area under the curve score of expression regulation by TFs, as estimated using SCENIC. B, Feature plots showing the distributions of active TFs and their targets in all malignant cells and in each malignant subgroup. UMAP plots of epithelial cells, color-coded for the expression of TFs (purple), for the area under the receiver operating characteristic curve of the estimated regulon activity of these TFs (red). C and D, Pseudo-time trajectory plots of all epithelial cell global transcriptomes by Monocle (C) and Slingshot (D). E, Differences in pathway activity (scored per cell by gene set variation analysis) in all malignant cell subclusters. F, Heat map showing expression level of all detected genes within chromosome region chr17q12 - chr17q21.2. G, The expression of selected genes (GRB7, KRT17, MIEN1, and RPL19) along the pseudo-time trajectory. H, Kaplan-Meier survival curve of selected gene expression using the median group cutoff showed the close relationship with patient overall survival probability in patients with CCA from The Cancer Genome Atlas data.