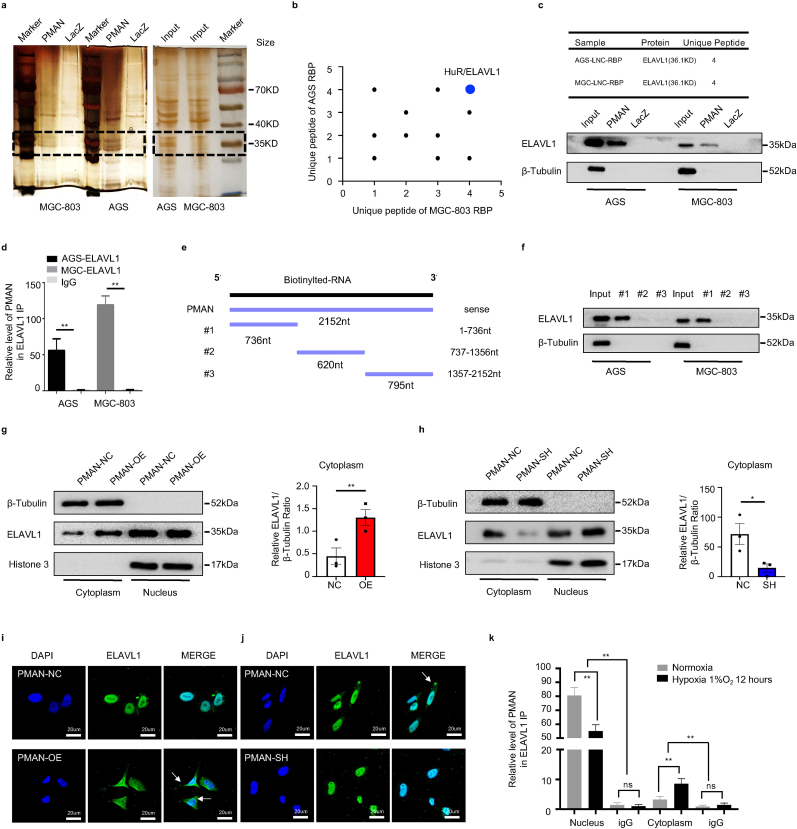
Fig. 5.
PMAN promotes ELAVL1 cytoplasmic translocation by direct bound to ELAVL1a RNA pull‐down proteins were precipitated in AGS cells and MGC-803 cells were detected by silver staining. b Proteins identification by LC-MS/MS in pulldowns of biotinylated PMAN from two independent experiences, two technical replicates. c Western blotting analysis of PMAN-binding protein ELAVL1 in AGS cells and MGC-803 cells. LacZ was treated as a negative control d RIP assay was performed with anti-ELAVL1 antibodies and control antibodies (IgG) in AGS cells and MGC-803 cells. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. e Three different PMAN deletion fragments were constructed based on the distribution of ELAVL1 specific binding sequence (GUUU). f The RNA binding protein ELAVL1 in three different PMAN deletion fragments were detected by western blotting in AGS cells and MGC-803 cells. g-h Protein nuclear-cytoplasmic fractionation assay showed the expression level of ELAVL1 in the subcellular fractions of PMAN-overexpression MGC-803 cells (g) and PMAN knockdown AGS cells (h) by western blotting and protein quantification on ImageJ. Histone H3 was treated as a nuclear control while β-tubulin was a cytoplasmic control. Mean ± SD is shown. Statistical analysis was conducted using Student's t-test. i-j IF showed ELAVL1 cytoplasmic fluorescence intensity (green) in PMAN-overexpression MGC-803 cells (i) and PMAN knockdown AGS cells (j). Scale bar, 20 μm k RIP assay was performed with anti-ELAVL1 antibodies and control antibodies (IgG) in cytoplasmic and nuclear components of MGC-803 cells during hypoxia and normoxia. Mean ± SD is shown. Statistical analysis was conducted using one-way ANOVA. Ns = nonsignificant (p > 0.05), *p < 0.05, **p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)