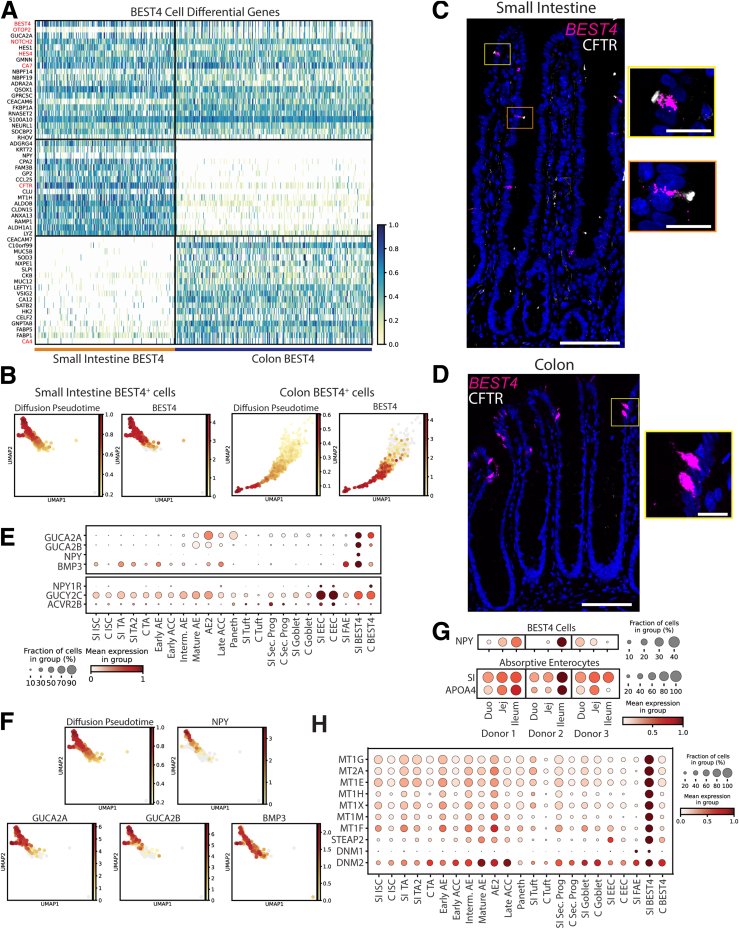
Figure 8.
BEST4+cells. (A) Heatmap of DEGs in BEST4+ cells vs other lineages (top; red: classic markers), SI vs colon BEST4+ cells (middle), colon vs SI BEST4+ cells (bottom). (B) UMAPs showing all SI (left) and colon (right) BEST4+ cells colored according to predicted diffusion pseudotime or expression of BEST4 mRNA. (C and D) In situ hybridization showing BEST4 mRNA (magenta), immunofluorescence staining CFTR protein (white), and nuclei (blue) in human (C) jejunum and (D) colon. Scale bars: 100 μm; 20 μm (zoomed panels). (E) Dotplot showing secreted genes (top) and their receptors (bottom) across lineages. (F) UMAPs of BEST4+ cells showing predicted diffusion pseudotime and expression of secreted peptides. (G) Expression of NPY, SI, and APOA1 across regions for each donor. (H) Dotplot showing genes involved in metal binding and endocytosis across lineages. UMAP, Uniform Manifold Approximation and Projection. CFTR, Cystic Fibrosis Transmembrane Conductance Regulator; GUCA2, Guanylate Cyclase Activator 2A; NPY, Neuropeptide Y.