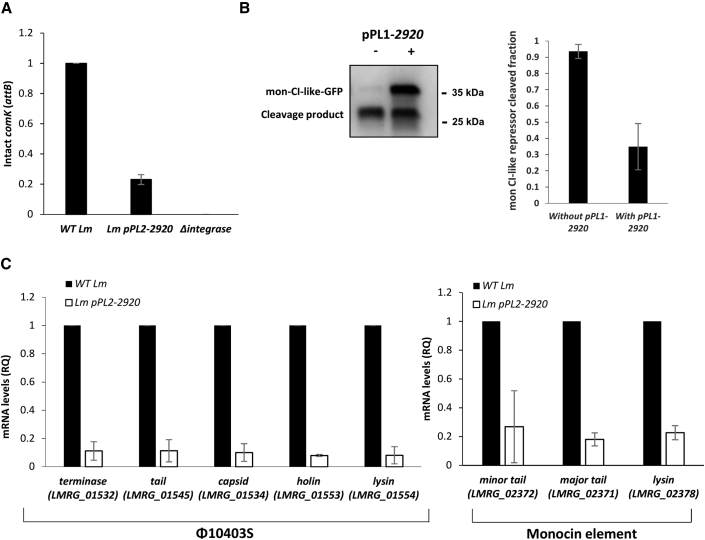
Figure 2.
LMRG_02920 inhibits the lytic induction of the phage and the mon element
(A) Quantitative real-time PCR analysis of the intact comK gene (representing φ10403S attB site) in WT Lm, Lm bacteria harboring pPL2-2920, and a mutant deleted of the φ10403S integrase gene (Δintegrase) as a control. Indicated strains were grown in BHI medium with MC treatment at 30°C for 3 h. The data are presented as relative quantity (RQ) relative to the levels in WT bacteria. The error bars represent the standard deviation of three independent experiments.
(B) Analysis of mon CI-like repressor cleavage by MpaR in the presence or absence of coexpression of LMRG_02920. Δφ/Δmon bacteria harboring a pPL2 plasmid expressing a translational fusion of mon CI-like-GFP under the Pconst promoter and MpaR under the PTetR promoter (pPL2-mon-cI-gfp-mpaR) were used in this experiment. This strain was also conjugated with the integrative pPL1 plasmid expressing LMRG_02920 under the PTetR promoter (pPL1-2920). To detect the in vivo cleavage of the mon CI-like repressor, equal amounts of total proteins from bacteria grown in the presence of MC were separated on 15% SDS-PAGE, blotted, and probed with anti-GFP antibody on western blots (a representative blot is shown in the left panel). Using ImageJ software, the amounts of the cleaved and non-cleaved mon CI-like repressors were quantified, and the mon CI-like cleaved fraction was calculated (right panel). The experiment was performed three times; error bars represent the standard deviation of three independent experiments.
(C) Quantitative real-time PCR analysis of select late-lytic genes of φ10403S (left panel) and the mon element (right panel) in WT Lm and Lm bacteria expressing LMRG_02920 (using pPL2-2920). Indicated strains were grown in BHI medium with MC treatment at 30°C for 3 h. mRNA levels are presented as RQ relative to their levels in WT bacteria. The error bars represent the standard deviation of three independent experiments.