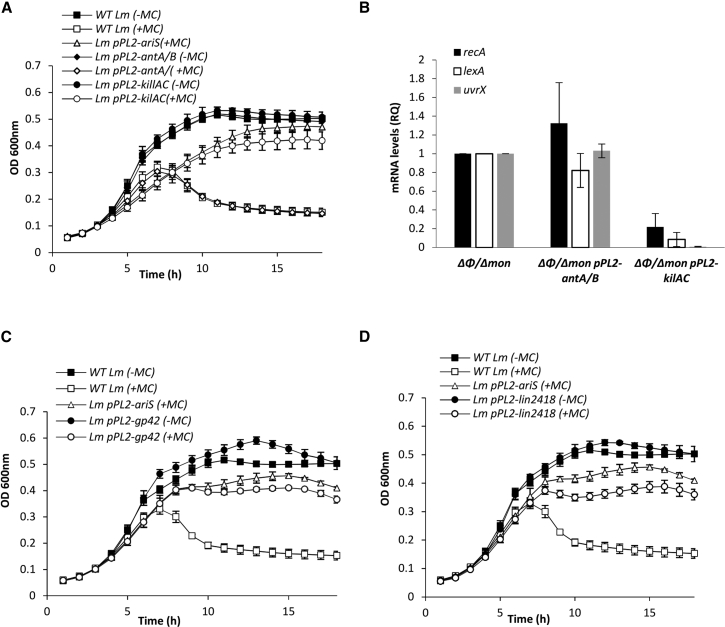
Figure 5.
The ANT/KilAC domain is responsible for the SOS inhibitory activity of AriS
(A) Growth analysis of WT Lm and Lm bacteria overexpressing LMRG_02920 (Lm pPL2-ariS [the same plasmid as pPL2-2920]), the N-terminal AntA/B domain (Lm pPL2-antA/B), or the C-terminal ANT/KilAC domain (Lm pPL2-kilAC). Bacteria were grown in BHI medium in the presence of anhydrotetracycline (AT) to induce the PTetR promoter and in the presence (+) or absence (-) of MC at 30°C. The error bars represent the standard deviation of three independent experiments and are sometimes hidden by the symbols.
(B) Quantitative real-time PCR analysis of representative SOS genes: recA, lexA, and uvrX (LMRG_02221) in Δφ/Δmon and Δφ/Δmon bacteria expressing each domain encoded by pPL2-antA/B and pPL2-kilAC plasmids. Indicated strains were grown in BHI medium with MC treatment at 30°C for 45 min. mRNA levels are presented as RQ relative to their levels in WT bacteria. The error bars represent the standard deviation of three independent experiments.
(C) Growth analysis of WT Lm with or without overexpression of Gp42 of the comK-phage of Lm WSLC1118 (Lm pPL2-gp42) in the presence (+) or absence (-) of MC at 30°C. Overexpression of ariS (Lm pPL2-ariS) was used as a control. The experiment was performed three times, and the figure shows a representative result. The error bars represent the standard deviation of three independent experiments and are sometimes hidden by the symbols.
(D) Growth analysis of WT Lm with or without overexpression of Lin2418 of the comK phage of Listeria innocua Clip 11262 (Lm pPL2-lin2418) in the presence (+) or absence (-) of MC at 30°C. Overexpression of ariS (Lm pPL2-ariS) was used as a control. The error bars represent the standard deviation of three independent experiments and are sometimes hidden by the symbols.