FIGURE 1.
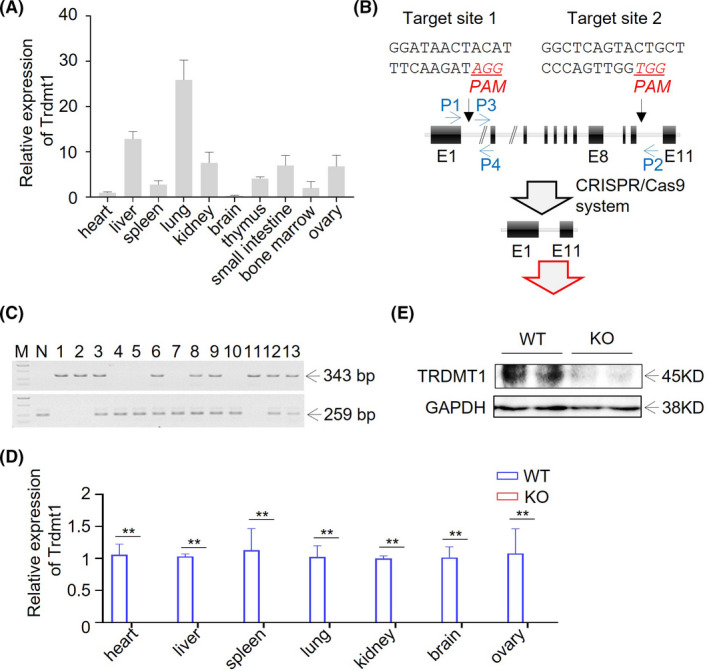
Generation of Trdmt1 gene knockout rat using CRISPR/Cas9. A, Relative expression of Trdmt1 in heart, liver, spleen, lung, kidney, brain, thymus, small intestine, bone marrow, and ovary tissues measured using real‐time PCR (n = 3). B, Schematic overview of CRISPR/Cas9‐mediated Trdmt1 gene deletion. The sgRNA targeting sites are shown. The protospacer adjacent motifs (PAMs) of sgRNA targeting sites are underlined in red and italic. Oligos used for sgRNAs preparation are listed in Table S1. Primers used for genotyping of Trdmt1 knockout rats are shown in blue and listed in Table S2. E, exon. C, PCR amplicon of Trdmt1 targeting locus. P1/P2 primer pair located outside of the sgRNA targeting sites and used for deleted DNA fragment detection. The P3/P4 primer pair located between the 2 sgRNA targeting sites and used for wild‐type DNA fragment detection. All primers used are listed in Table S2. M, marker for DL2, 000 (TaKaRa). N, negative control (wild type). D, The relative expression of Trdmt1 in heart, liver, spleen, lung, kidney, brain, and ovary tissues confirmed in Trdmt1 knockout rats using real‐time PCR (n = 3). E, Protein level of TRDMT1 in lung of Trdmt1 knockout rats. WT, wild‐type rats; KO, Trdmt1 knockout rats. Mean ± SEM; *P < 0.05, **P < 0.01
