Summary
Clinical trials that tested the antitumor activity of γδ T cells have been mostly unsuccessful. To address this, we expanded human Vγ9Vδ2 T cells in TGFβ1, a cytokine which enhances their viability, trafficking properties, and intrinsic antitumor activity. This protocol summarizes the production and in vitro functional characterization of TGFβ1 educated human Vγ9Vδ2 cells and highlights their compatibility with chimeric antigen receptor (CAR) engineering. We also describe in vivo testing of the antitumor activity of these CAR T cells in mice.
For complete details on the use and execution of this protocol, please refer to Beatson et al. (2021).
Subject areas: Cell Biology, Cell culture, Flow Cytometry/Mass Cytometry, Cell-based Assays, Cancer, Health Sciences, Immunology, Model Organisms, Molecular Biology, Biotechnology and bioengineering
Graphical abstract

Highlights
-
•
TGFβ promotes viability and retards differentiation of Vγ9Vδ2 cells
-
•
TGFβ-educated Vγ9Vδ2 cells achieve greater invasion and anticancer activity
-
•
Specificity of these cells can be reprogrammed using chimeric antigen receptors
Clinical trials that tested the antitumor activity of γδ T cells have been mostly unsuccessful. To address this, we expanded human Vγ9Vδ2 T cells in TGFβ1, a cytokine which enhances their viability, trafficking properties, and intrinsic antitumor activity. This protocol summarizes the production and in vitro functional characterization of TGFβ1 educated human Vγ9Vδ2 cells and highlights their compatibility with chimeric antigen receptor (CAR) engineering. We also describe in vivo testing of the antitumor activity of these CAR T cells in mice.
Before you begin
Experimental consideration
-
1.
γδ T cells play a key role in immune surveillance for malignant transformation (Kabelitz et al., 2020). Moreover, the presence of intra-tumoral γδ T cells is strongly predictive of improved prognosis across a range of cancer types (Gentles et al., 2015). The dominant circulating γδ T cell subtype (δ2 subtype) expresses a Vγ9Vδ2 T cell receptor that detects phosphoantigen intermediates of mevalonate metabolism, a metabolic pathway that is upregulated in transformed cells. These cells can be selectively activated using aminobisphosphonate drugs such as zoledronic acid [ZOL]. Non δ2 γδ T cells are also present in the circulation, albeit at lower levels. These cells also possess intrinsic anti-tumor activity. In order to activate both γδ T cell subtypes, an immobilized pan-γδ T cell receptor antibody may be used. Given the intrinsic anti-tumor activity of both δ2 and non δ2 γδ T cells and the fact that neither subtype causes graft versus host disease, these cells are the subject of great interest as a potential off-the-shelf approach to adoptive cancer immunotherapy.
-
2.
Recombinant human TGFβ1 available from different suppliers varies in its bioactivity. We use TGFβ1 from Bio-Techne and have validated its activity in house using a TGFβ1 reporter system (Abe et al., 1994). We have tested products from other manufacturers, although none have performed as robustly.
-
3.
We have used either ZOL or an immobilized anti-γδ TCR antibody (11F2 or B1 clones) to elicit the activation of circulating γδ T cells, enabling their subsequent cytokine-driven expansion. Using the immobilized anti-γδ TCR antibody method, non-Vδ2 cells are also expanded. We have not tested other antibodies for the expansion of these cells (e.g., anti-Vγ9 antibodies). Moreover, we have not tested this method for the expansion of tissue-resident γδ T cells or using other isoforms of TGFβ.
-
4.
Extensive optimization of the system was carried out using different TGFβ1 concentrations, dosing regimens and media. Importantly, these effects of TGFβ1 could be recapitulated in media containing human serum, and in other serum-free media. However, the serum-free protocols described in this paper were found to be optimal.
-
5.
A variety of different starting materials were trialed as a source of Vγ9Vδ2 cells with freshly isolated whole blood providing the most consistent and optimal expansion numbers. Other starting materials tested were leukocytes cones, whole blood from the National Blood and Tissue Service (Tooting, London) and frozen PBMCs.
-
6.
Except for centrifugation and the blood draw, all steps in this protocol involving cell culture are performed in a laminar flow hood using aseptic technique.
Institutional permissions
All in vivo experimentation must comply with local regulatory requirements. In this case, experimental work was compliant with U.K. Home Office guidelines, as specified in project license number P23115EBF. In addition, all work was approved by the King’s College London animal welfare and ethical review body (AWERB).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human/primate EGF biotinylated – 1/150 dilution | R&D Systems | Cat# BAF236; RRID: AB_356307 |
| CD27 – PE (M-T271) – 1/100 dilution |
BioLegend UK | Cat# 356405, RRID: AB_2561824 |
| CD45RA – BV605/ APC (HI100) – 1/100 dilution |
BioLegend UK | Cat# 304150, RRID: AB_2564158 |
| PE Goat anti-mouse IgG – 1/100 dilution |
BioLegend UK | Cat# 405307 |
| Streptavidin PE (SA-PE) - – 1/150 dilution | BioLegend UK | Cat# 405203 |
| TCR Pan γδ purified (11F2) – coat plates with 0.8 mg/mL | BD Biosciences | Cat# 347900, RRID: AB_400356 |
| TCR Pan γδ - FITC (B1) – 1/100 dilution |
BD Biosciences | Cat# 559878, RRID: AB_397353 |
| Mouse IgG1 FITC isotype – 1/100 dilution |
BD Biosciences | Cat# 345815, RRID: AB_2868833 |
| Ultra-LEAF CD11a blocking antibody – block with 10 μg/mL | BioLegend UK | Cat# 301233, RRID: AB_2832576 |
| Ultra-LEAF Mouse IgG1 LEAF – block with 10 μg/mL | BioLegend UK | Cat# 400166, No RRID available |
| Ultra-LEAF Pan γδ TCR (B1) - coat plates with 0.8 mg/mL | BioLegend UK | Cat# 331235, RRID: AB_2814201 |
| Bacterial and virus strains | ||
| DH5α E. coli | New England Biolabs | Cat# c2987h |
| SFG retroviral vector | Dr Michel Sadelain, Memorial Sloan Kettering Cancer Center | N/A |
| Biological samples | ||
| Human blood | Healthy volunteers, male and female, aged 18–65 years | Approved by the West of Scotland Research Ethics Committee 3 (REC reference 18/WS/0047) |
| Chemicals, peptides, and recombinant proteins | ||
| Antibiotic antimycotic (100×) | Thermo Fisher Scientific | Cat# 15240062 |
| Bovine serum albumin | Sigma-Aldrich | Cat# A9418 |
| D-luciferin | Cambridge Bioscience | Cat# B3000-1G |
| DMEM medium | Lonza | Cat# BE12-604Q |
| Disodium edetate (EDTA) | Sigma-Aldrich | Cat# BP1224 |
| Engelbreth-Holm-Swarm (EHS) basement membrane extract (BME) | Sigma-Aldrich | Cat# 126-2.5 |
| Fetal bovine serum | Gibco | Cat# 26140079 |
| Ficoll®-Paque PLUS | GE Healthcare | Cat# GE17-1440-02 |
| GeneJuice® transfection reagent | Sigma-Aldrich | Cat# 70967-3 |
| Isoflurane | Sigma-Aldrich | Cat# 792632 |
| L-Glutamine solution | Sigma-Aldrich | G7513-100ML |
| Luciferin | Regis Technologies | Cat# 115144-35-9 |
| MTT | Apollo Scientific | Cat# BID2165 |
| Pamidronate sodium | Pfizer Ltd. | N/A |
| PBS | Sigma-Aldrich | D8537 |
| Proleukin (aldesleukin), human recombinant interleukin (IL)-2 | Clinigen Group | N/A |
| Recombinant Human TGF-β1 | Bio-Techne | Cat# 240-B |
| RetroNectin® recombinant human fibronectin fragment | Takara Bio | Cat# T100B |
| Sodium Citrate | USP | 1613859 |
| TexMACSTM medium | Miltenyi Biotec | 130-097-196 |
| Trypan Blue | Sigma-Aldrich | Cat# T8154 |
| Zometa | Novartis | N/A |
| Critical commercial assays | ||
| Human uncoated IL-2 ELISA kit | Life Technologies Ltd | Cat# 88-7025-88 |
| Human IFN-gamma DUOset ELISA | Bio-Techne | Cat# DY285B |
| Experimental models: Cell lines | ||
| BT-20 | Breast Cancer Now Research Unit, King’s College London | ATCC Cat# HTB-19 RRID CVCL_0178 |
| BxPC-3 | Barts Cancer Institute, Queen Mary University of London | ATCC Cat# CRL-1687 RRID CVCL_0186 |
| HEK293T | ATCC | Cat# CRL-3216 RRID: CVCL_0063 |
| MDA-MB-231 | Breast Cancer Now Research Unit, King’s College London | ATCC Cat# HTB-26 RRID: CVCL_0062 |
| MDA-MB-468 | Breast Cancer Now Research Unit, King’s College London | ATCC Cat# HTB-132 RRID CVCL_0419 |
| Ovsaho | Japanese Collection of Research Bioresources Cell Bank | Cat# JCRB1046 RRID CVCL_3114 |
| PG13 | European Collection of Authenticated Cell Culture | Cat# 95110215 RRID CVCL_4273 |
| TOV-21G | Prof Sadaf Ghaem-Maghami, Imperial College London | ATCC Cat# CRL-11730 RRID: CVCL_3613 |
| Experimental models: Organisms/strains | ||
| NOD.Cg-PrkdcSCID Il2rgtm1Wjl/SzJ (NSG) mice, male and female, aged 6–10 weeks | Charles River | Strain code: 614 |
| Recombinant DNA | ||
| PeqPam plasmid | Gift of Dr M Pule, University College London | N/A |
| pCAR-H/T | CAR Mechanics Group, King’s College London | (Muliaditan et al., 2021) |
| RDF plasmid | Gift of Prof M Collins, University College London | N/A |
| Software and algorithms | ||
| FlowJo v.10 Software | Tree Star | https://www.flowjo.com/ |
| Gene Designer | DNA2.0 | https://www.atum.bio/resources/tools/gene-designer |
| Prism 9 | GraphPad | https://www.graphpad.com/scientificsoftware/prism |
| SnapGene | GSL Biotech | https://www.snapgene.com/ |
| Other | ||
| Adhesive PCR Plate Seals | Thermo Fisher Scientific | Cat# AB0558 |
| MicroAmp™ Optical 96-Well Reaction Plate | Thermo Fisher Scientific | Cat# n8010560 |
| RTCA E-plates | Agilent | Cat# 300601010 |
| Sodium heparin blood collection tubes | BD Biosciences | Cat# 367874 |
| ThinCertTM inserts for 24 well plates (6.5 mm, 3 μm pore size) | Greiner Bio-One | Cat# 662631 |
| xCELLigence RTCA MP | Agilent | N/A |
| BD LSRFortessaTM X-20 | BD Biosciences | N/A |
Materials and equipment
TexMACSTM+ medium
| Reagent | Final concentration | Amount |
|---|---|---|
| TexMACSTM medium | ×1 | 490 mL |
| L-Glutamine (×100) | 2 mM | 5 mL |
| Antibiotic Antimycotic (×100) | ×1 | 5 mL |
| Total | N/A | 500 mL |
Note: Store TexMACSTM+ medium at 4°C for up to 2 weeks.
D10 medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM medium | ×1 | 440 mL |
| Fetal bovine serum | 10% | 50 mL |
| L-Glutamine (×100) | 2 mM | 5 mL |
| Antibiotic Antimycotic (×100) | ×1 | 5 mL |
| Total | N/A | 500 mL |
Note: Store D10 medium at 4°C for up to 2 weeks.
FACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | ×1 | 99 mL |
| Disodium edetate (0.5 M stock) | 0.5 mM | 100 μL |
| Fetal bovine serum | 1% | 1 mL |
| Total | N/A | 100 mL |
Note: Store Disodium edetate (0.5 M stock) for up to 1 year at 4°C.
Note: Prepare FACS buffer fresh before use.
Equipment
| Flow cytometer | Analytical software | Version |
|---|---|---|
| BD LSRFortessaTM X-20 | FlowJoTM | V10.8.1 |
Step-by-step method details
Timing: 15 or more days, depending on expansion methodology used
Expansion of activated Vγ9Vδ2 T cells in the presence of TGFβ1.
This step describes methods used to activate and expand Vγ9Vδ2 T cells over a 2 week period in serum-free medium supplemented with IL-2 and TGF-β1.
Note: Cells expanded using the combination of IL-2 and TGF-β1 are designated γδ[T2] cells. If cells are expanded in the same way but using IL-2 alone, they are referred to as γδ[2] cells.
Note: Our RNA sequencing data suggests that cells are not phenotypically mature at day 8 (Beatson et al., 2021). We therefore recommend completing the 15 day culture period.
Day -1.
-
1.
Alternative (antibody activation method only): Coat 24, 12 or 6 well plates (depending on the numbers of PBMCs expected) overnight (12–16 h) at 4°C with 0.8 mg/mL pan γδ TCR antibody (11F2 or B1 clones) contained in a volume of 1 mL, 2 mL or 3 mL PBS, respectively.
CRITICAL: We have not tested other pan γδ TCR antibodies for their ability to stimulate the activation of these cells.
Day 0.
-
2.
Withdraw blood (typically 45 mL) from a consenting healthy volunteer and mix with 5 mL sodium citrate in pre-prepared 50 mL Falcon tubes (stored up to 6 months at 4°C).
Note: Alternatively, blood can be collected into sodium heparin blood collection tubes.
-
3.
Gently mix blood with an equal volume of PBS (1:1 ratio).
-
4.
Gently layer the blood/PBS mix (using the gravity setting on a pipette controller) on top of 15 mL Ficoll Paque Plus in 50 mL Falcon tubes.
-
5.
Centrifuge the 50 mL tubes at 750 × g for 30 min at 20°C (acceleration and brake set to 0) to separate the PBMC cell fraction.
-
6.
Transfer the PBMC layer, seen as an interface between the plasma and the Ficoll layer, using a sterile Pasteur pipette to a fresh 50 mL Falcon tube.
Note: After centrifugation, the layer above the PBMC fraction which contains plasma can be carefully removed using a Pasteur pipette to make the PBMC layer easier to harvest.
-
7.
In the 50 mL tube containing the PBMCs, add PBS to a final volume of 50 mL and centrifuge at 500 × g for 5 min. The acceleration and brake can be set back to 9.
-
8.
Aspirate and discard the supernatant leaving the pelleted cells.
-
9.
If more than one tube has been used (due to the volume size), resuspend the pellets in TexMACSTM+ and combine in a single tube. Bring the cells up to a final volume of 50 mL of PBS for a second wash.
-
10.
Centrifuge at 500 × g for 5 min.
-
11.
Aspirate and discard the supernatant.
-
12.
Resuspend the cell pellet an appropriate volume of TexMACSTM+ medium (see materials and equipment Table above; usually 10 mL per 45 mL blood).
CRITICAL: We have not tested other serum free media for their ability to support the expansion of γδ T-cells.
-
13.
Count live cells using trypan blue staining and a hemocytometer. Resuspend cells at 3 × 106/mL.
Note: Samples are normally diluted 1 in 10 with PBS prior to cell counting.
-
14.
Alternative (antibody activation method only – to expand both δ2 and non-δ2 γδ T cells): Aspirate the antibody-containing PBS solution from plates set up in step 1. Wash each well once with PBS. Plate PBMC immediately at a density of 3 × 106 cells/mL in 4 mL TexMACSTM+ medium per well of a 6 well plate. Add recombinant IL-2 (100 U/mL) and recombinant TGF-β1 (5 ng/mL). After 72 h, remove the cells are removed from this plate.
OR
-
15.
Alternative (zoledronic acid method only – to expand δ2 γδ T cells): Plate PBMC at a density of 3 × 106 cells/mL in TexMACSTM+ medium. Add zoledronic acid (1 mg/mL), together with recombinant IL-2 (100 U/mL) and recombinant TGF-β1 (5 ng/mL).
Note: We do not observe consistent differences in total yield of γδ T cells achieved using the two methods although the relative proportion of non δ2 γδ T cells that expand is greater using the antibody method.
Days 1–14.
-
16.Monitor cell density by microscopy and cell counting every 2–3 days.
-
a.If cell density exceeds 1 × 106 cells/mL, add 100% TexMACSTM+ medium followed by recombinant IL-2 (100 U/mL) and recombinant TGF-β1 (5 ng/mL).
-
b.If cell density is below this level, add cytokines alone.
-
a.
Note: Cytokine addition is corrected to the total volume of medium present. Where necessary, split and expand cultures into additional wells on the chosen plate type, and thereafter into T75 flasks.
Day 15.
-
17.Determine the purity of γδ T-cells by flow cytometry:
-
a.Transfer a minimum of 5 × 104 γδ[2] and γδ[T2] cells into two separate 5 mL flow cytometry tubes.
-
b.Wash each tube with 2 mL FACS buffer (PBS with 0.5 mM EDTA and 1% FCS) and centrifuge at 400 × g for 5 min.
-
c.Discard supernatant and resuspend the cell pellet in 50 μL PBS.
-
d.Add 5 μL anti-human γδ TCR-FITC antibody (BD Biosciences; clone B1; Cat# 559878, RRID: AB_397353 – see key resources table) to one tube containing γδ[2] and γδ[T2] cells.Note: Other anti-human γδ TCR antibodies suitable for flow cytometry are commercially available and may potentially be used for this step.
-
e.Add 5 μL FITC-conjugated isotype control to the second tube containing γδ[2] and γδ[T2] cells. Place tubes on ice for 30 min.
-
f.Wash each tube with 2 mL FACS buffer and centrifuge at 400 × g for 5 min.
-
g.Discard supernatant and resuspend cells in 100–250 μL PBS.
-
h.Analyze using flow cytometry.
 Pause Point: Cells can be maintained in culture for up to day 21. In this case, continue the feeding regimen as described in step 16.Note: Using this method, a median purity of 92.7% γδ T cells is achieved. The lowest purity achieved at which acceptable anti-tumor activity was observed was 74.9%. Flow sorting can be used to improve purity and also to remove contaminating αβ T cells and B cells, which are undesirable impurities in the event that allogeneic γδ T cells are infused for therapeutic purposes.Note:Figure 1 presents an example in which γδ[2] and γδ[T2] cells were stained with differentiation markers, CD27 and CD45RA (Dieli et al., 2003). γδ[T2] cultures generally exhibit greater viability and display a less differentiated phenotype (Beatson et al., 2021).Note: The evaluation assays described in the sections that follow are performed independently of each other, rather than representing a sequentially timed protocol.
Pause Point: Cells can be maintained in culture for up to day 21. In this case, continue the feeding regimen as described in step 16.Note: Using this method, a median purity of 92.7% γδ T cells is achieved. The lowest purity achieved at which acceptable anti-tumor activity was observed was 74.9%. Flow sorting can be used to improve purity and also to remove contaminating αβ T cells and B cells, which are undesirable impurities in the event that allogeneic γδ T cells are infused for therapeutic purposes.Note:Figure 1 presents an example in which γδ[2] and γδ[T2] cells were stained with differentiation markers, CD27 and CD45RA (Dieli et al., 2003). γδ[T2] cultures generally exhibit greater viability and display a less differentiated phenotype (Beatson et al., 2021).Note: The evaluation assays described in the sections that follow are performed independently of each other, rather than representing a sequentially timed protocol.
-
a.
Figure 1.
Assessment of differentiation state of γδ[T2] and γδ[2] cells
Cells were co-stained with anti-γδ TCR-FITC, CD27-PE and CD45RA-APC antibodies. Quadrant settings were determined using isotype controls. Note the less differentiated nature of γδ[T2] cells. CM – central memory; EM- effector memory; Term – terminally differentiated cells.
Evaluation of in vitro real time tumor cytolytic activity of γδ[T2] cells
Timing: Up to 6 days
This step describes methods used to quantify the tumor cell killing activity of TGF-β-educated Vγ9Vδ2 T cells using the xCELLigence MP impedance analyzer.
Note: It is recommended to use tumor cell lines that have been passaged 20 times or less.
-
18.
Plate MDA-MB-231 tumor cells (2 × 104 cells each) on a 96 well electronic microtiter plate (Agilent; Cat# 300601010) in 200 μL D10 medium.
Note: DMEM medium is available from many commercial suppliers and may be used for this step.
-
19.
After 24 h, pulse the cells with ZOL (3 μg/mL) or medium alone as a control.
-
20.
After a further 24 h, remove 100 μL medium from each well. Add γδ[T2] and γδ[2] cells in 100 μL TexMACSTM+ medium to achieve a final 1:1 E:T ratio.
Note: Add cells gently to minimize disruption to tumor monolayers.
-
21.
Perform dynamic monitoring of adherent tumor viability/ proliferation using an xCELLigence MP impedance analyzer.
Note: This equipment measures cellular impedance (and thereby cellular viability) in a label-free manner. The xCELLigence MP impedance analyzer can host up to six 96-well electronic microplates. The analyzer is maintained in an incubator (37°C, 5% CO2).
-
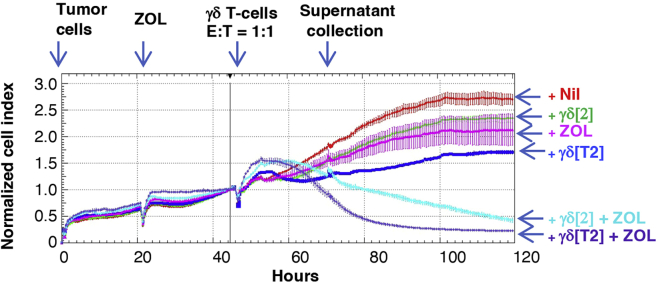
22.
A representative example of the analysis is shown in Figure 2.
Figure 2.
Real time cytolytic activity of γδ[T2] and γδ[2] cells
In this example, MDA-MB-231 tumor cells were added to a 96 well electronic microtiter plate. Zoledronic acid (3 μg/mL) and either γδ[2] or γδ[T2] cells were added after 24 and 48 h respectively. Tumor cell number is normalized to the number present at the time of γδ T cell addition (“normalized cell index”).
Evaluation of in vitro tumor cytolytic activity and cytokine release by γδ[T2] cells
Timing: 3–5 days
This step describes methods used to quantify the tumor cell killing activity of TGF-β-educated Vγ9Vδ2 T cells using an MTT assay. Measurement of cytokine release by these cells by ELISA is also described.
Note: It is recommended to use tumor cell lines that have been passaged 20 times or less.
-
23.
Plate tumor cells (1 × 105 cells each) in a 24 well microtiter plate in 1 mL D10 medium (see materials and equipment Table above).
-
24.
After 24 h, pulse tumor cell monolayers with the indicated concentration of ZOL or pamidronic acid (PAM).
Note: These are alternative aminobisphosphonate drugs that cause phosphoantigen accumulation in tumor cells, thereby sensitizing them to γδ T cells.
-
25.
After a further 24 h, remove half of the medium and add γδ[T2] and γδ[2] cells in 500 μL TexMACSTM+ medium to achieve a final 5:1 E:T ratio.
-
26.
After a further 24 h, collect 200 μL supernatant from each well for cytokine analysis by ELISA, as described by the manufacturers.
Note: Supernatants can be harvested 24–72 h after addition of γδ T cells. While IFN-γ is readily detectable at these time points, IL-2 levels are much lower and are optimally detected at 24 h after γδ T cell addition.
Note: Supernatants are diluted 1 in 5 to measure IFN-γ and are analyzed undiluted to measure IL-2. Follow ELISA manufacturer’s protocols to measure IFN-γ and IL-2 for this step.
Note:Figure 3 provides examples of IFN-γ and IL-2 release by γδ[T2] and γδ[2] cells when co-cultivated with a panel of triple negative breast cancer cell lines that had been pre-sensitized with the indicated concentration of PAM or ZOL. Further examples are provided in (Beatson et al., 2021).
-
27.Seventy two hours after addition of γδ[T2] and γδ[2] cells, assess residual tumor cell viability using the 3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (MTT) assay.
-
a.Prepare MTT to a concentration of 500 μg/mL in D10 medium (13 mL/24 well plate).
-
b.Carefully remove media from each well of one 24 well plate (leaving two plates for 48 h and 72-h analysis).
-
c.Pipette 500 μL of 500 μg/mL MTT solution into each well.
-
d.Incubate at 37°C & 5% CO2 for 1 h.
-
e.Aspirate MTT solution from each well being careful not to disturb the monolayers.
-
f.Add 500 μL DMSO into each well.
-
g.Gently swirl the plate to solubilize the formazan crystals.
-
h.Read absorbance at 560 nm using a plate reader.
-
i.Calculate residual tumor cell viability using the following equation:
-
a.
Note:Figure 4 provides examples of tumor cell killing by γδ[T2] and γδ[2] cells when co-cultivated with a panel of ovarian cancer cell lines that had been pre-sensitized with the indicated concentration of PAM or ZOL. Further examples involving triple negative breast cancer cell lines are provided in (Beatson et al., 2021).
Figure 3.
Cytokine release by γδ[T2] and γδ[2] cells
γδ[2] cells and γδ[T2] cells were co-cultivated with the indicated triple negative breast cancer monolayers (5:1 E:T ratio) that had been pulsed 24 h earlier with ZOL or PAM at concentrations specified. Release of IFN-γ and IL-2 was measured by ELISA in supernatants collected after 24 h. All data show mean + SD of n=3–5 replicates. All statistical analysis was performed using two-way ANOVA, making comparison between γδ[2] cells and γδ[T2] cells. ∗ p<0.05; ∗∗ p<0.01; ∗∗ p<0.001;∗∗∗∗ p<0.0001; NS – not significant.
Figure 4.
Tumor cytolytic activity of γδ[T2] and γδ[2] cells
γδ[2] cells and γδ[T2] cells were co-cultivated with the indicated ovarian cancer monolayers (5:1 E:T ratio) that had been pulsed 24 h earlier with ZOL (at concentrations specified). Black lines represent monolayers without the addition of γδ cells. Residual tumor cell viability was measured after 72 h by MTT assay. All data show mean + SD of n=3–6 replicates. All statistical analysis was performed using two-way ANOVA, making comparison between γδ[2] cells and γδ[T2] cells. NS – not significant.
Evaluation of basement membrane invasion by γδ[T2] cells
Timing: 3 days
This step describes methods used to quantify the ability of TGF-β-educated Vγ9Vδ2 T cells to invade basement membrane extract.
Day -1.
-
28.
Block the required number of wells in rows B and C of a 24 well plate(s) with 1% BSA in PBS overnight (12–16 h) at 4°C.
-
29.
Place sterile 1.5 mL Eppendorfs, 20 and 200 μL pipette tips at 4°C overnight (12–16 h).
Day 0.
-
30.
Thaw Engelbreth-Holm-Swarm (EHS) mouse sarcoma-based basement membrane extract (BME) on ice.
CRITICAL: An icebox should be placed within a tissue culture hood to maintain sterility and low-temperature.
-
31.
Aspirate the blocking solution (see step 1) from the 24 well plate and replace with 650 μL TexMACSTM+ medium per well.
-
32.
Mix BME with cold TexMACSTM+ media at a 1:1 ratio on ice (within hood) using a cold 1.5 mL Eppendorf and pipette tips (keep on ice).
Note: The volume prepared should allow for coating of the required number of Transwell inserts with 80 μL of the mixture. A small excess should be prepared to allow for losses during pipetting.
-
33.
Coat ThinCertTM inserts (6.5 mm, 3 μm pore size) with 80 μL BME/TexMACSTM+ solution using cold pipette tips and placed in rows A and D of the 24 well plate.
CRITICAL: Make sure the solution is spread evenly and there are no bubbles. If there are bubbles, very gently use a tip to remove, being careful not to touch the membrane of the insert. If this is not possible, take a new insert and start again.
-
34.
Place the 24 well plate in an incubator (37°C) for at least 30 min or at room temperature (approximately 20°C–22°C) for >1 h to allow the BME to solidify.
-
35.
Remove expanded γδ[T2] and γδ[2] cells from culture into a 15 mL Falcon tube and count live cells using trypan blue and a hemocytometer.
Note: there is no need for trypsin or any disassociation agent as the cells do not attach to plastic.
-
36.
Centrifuge expanded γδ[T2] and γδ[2] cells at 350 × g for 5 min at room temperature (approximately 20°C–22°C). Remove supernatant and resuspend the cells at a density of 1 × 106/mL.
-
37.
If antagonism of cell surface proteins is being assessed, treat γδ T cells with an appropriate blocking antibody or control in PBS prior to the start of the assay.
Note: In the example shown in Figure 5, the effect of CD11a blockade has been tested, making comparison with an isotype control at the same concentration. In this example, γδ T cells are treated with 10 μg/mL of anti-CD11a or isotype control immediately prior to placement of cells in ThinCertTM.
-
38.
If migration towards a particular factor or supernatant is being assayed, add the relevant factor to (or substitute for) medium in the appropriate well in rows B-C. This has not been tested in the example shown in Figure 5.
Note: Steps 37 or 38 should be performed <5 min before the γδ[T2] and γδ[2] cells are added to coated ThinCertTM inserts in rows A and D.
-
39.
Remove the 24 well culture plate from the incubator.
-
40.
Gently add 150 μL of re-suspended γδ[T2] and γδ[2] cells (e.g., 1.5 × 105 cells) to the top of the coated ThinCertTM inserts in rows A and D, thereby placing the cells on top of the BME.
-
41.
The assay commences when these inserts are transferred into the media-containing wells in rows B and C.
-
42.
At predetermined time points, carefully remove the inserts from the media and place these in the corresponding columns in rows A and D of the 24 well plate.
-
43.
Count the number of cells present in the medium using a hemocytometer. It is advisable to take three measurements for each well, as there can be large variations.
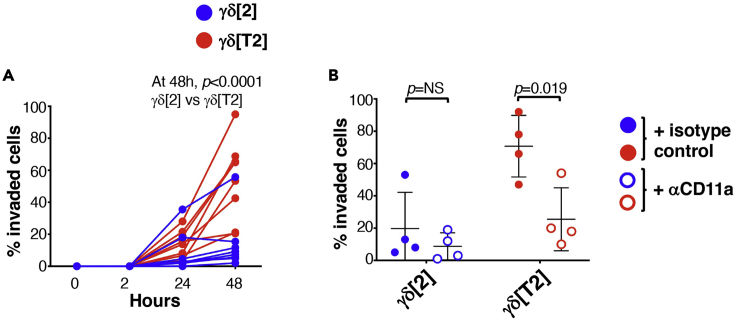
Note: A representative example of this protocol is shown in Figure 5. Comparison is made between γδ[T2] and γδ[2] cells. Inhibition of invasion by CD11a blockade has also been evaluated, implicating CD11a in this process.
Figure 5.
Basement membrane extract invasion by γδ[T2] and γδ[2] cells
(A) γδ[2] or γδ[T2] cells were placed on BME within ThinCertsTM which were placed in TexMACSTM+. Cells that invaded into the lower well were enumerated at the indicated time points (n=8 per group).
(B) The experiment in A was repeated in the presence of a blocking antibody directed against CD11a or an isotype control (mean +/- SD, n=4). Invaded cells (%) were measured at 48 h. Statistical analysis was performed using an unpaired Student’s t-test.
Engineering of γδ[T2] T cells to express a chimeric antigen receptor
Timing: Minimum 2 weeks (to allow for transduction and expansion of the cells)
This step describes methods used to engineer TGF-β-educated Vγ9Vδ2 T cells to express a chimeric antigen receptor.
Note: For vector design protocols and transient retroviral production protocols using triple transfected 293T cells, please see Larcombe-Young et al. (currently under review as a Star Protocol). Using this method, viral titer typically ranges from 5–10 × 105 viral particles/mL. Viral titer is not routinely determined prior to proceeding as described below.
Note: Parallel (p)CAR is a recently described technology in which a second generation (CD28-containing) CAR is co-expressed with a 4-1BB containing chimeric co-stimulatory receptor. The pCAR-H/T parallel CAR co-targets MUC1 and ErbB dimers and is shown in schematic form in Figure 6. Transduced cells are conveniently detected using anti-EGF antibody which binds specifically to one targeting moiety present in pCAR-H/T. Further details of the pCAR-H/T pCAR are provided in (Muliaditan et al., 2021).
Note: this protocol is optimized for transduction of γδ[T2] cell cultures which have been cultured for 72 h on immobilized pan γδ TCR antibody (11F2 clone).
-
44.24 h before transduction:
-
a.Add 100 IU/mL recombinant human IL-2 to activated γδ[T2] cells.
-
b.Add 200 μL RetroNectin® (1 mg/mL stock solution) to 18 mL sterile PBS in a 50 mL Falcon tube.
-
c.Using a Pasteur pipette, aliquot 3 mL into each well of a non-tissue culture-treated 6 well plate.
-
d.Wrap the 6 well plate in Saran Wrap and store overnight (12–16 h) in a refrigerator at 4°C.
-
a.
Note: 1 million cells are transduced in each well of the RetroNectin®-coated 6 well plate. Calculate how many wells are needed for each construct to be transduced.
CRITICAL: RetroNectin® can stick to certain plastics. Use polypropylene Pasteur pipettes to minimize protein loss during reagent preparation.
-
45.On the day of transduction:
-
a.Rapidly thaw 1.5 mL aliquots of viral supernatant in a 37°C water bath for the retroviral constructs of choice.Note: A total of 4.5 mL viral supernatant is required per 1 × 106 cells to be transduced, of which 1.5 mL will be used to pre-load the appropriate well of the RetroNectin®-coated 6 well plate.
-
b.Spray cryovials containing viral supernatant with 70% ethanol and transfer into a laminar flow hood.
-
c.Pre-load the RetroNectin®-coated 6 well plate with viral vector by removal of RetroNectin® from each individual well and prompt addition (to avoid drying) of 1.5 mL of thawed viral supernatant.
-
d.Incubate virus coated six well plate for 2 h or overnight (12–16 h) at 4°C.
-
e.Retrieve activated γδ[T2] cells from antibody-coated plate and transfer into a Falcon tube. After gentle mixing by inversion, count using a hemocytometer and Trypan blue.
-
f.Aspirate the 1.5 mL viral vector from the preloaded RetroNectin®-coated 6 well plate.
-
g.Add 3 mL of freshly thawed viral supernatant to each well.
-
h.Add 1 × 106 activated γδ[T2] cells per well of the 6 well plate.
-
i.For non-transduced control γδ[T2] cells, add 3 mL D10 medium.
-
j.Add IL-2 to each well to a final concentration of 100 IU/mL.
-
k.Incubate cells at 37°C & 5% CO2 for 72 h.Note: Activated T-cells should not be washed prior to counting and transduction as this will remove potentially stimulatory factors present in the medium that they have conditioned.Note: Do not use all activated T-cells for transduction. Untransduced T-cells will later be required, for example, as a control for flow cytometry analysis and/ or functional studies.
-
a.
-
46.
Every 2 days (3 days if leaving over weekends), feed γδ[T2] cells with 100% volume increase of TexMACSTM+ medium, with the addition of recombinant human IL-2 to a final concentration of 100 IU/mL and recombinant human TGFβ1 to 5 ng/mL. Over this interval, maintain T cells at 37°C and 5% CO2.
Note: If cell density is below 5 × 105/mL, add 100 IU/mL recombinant human IL-2, and 5 ng/mL recombinant human TGFβ1 only.
-
47.On day 15 of the culture, determine the transduction efficiency of the γδ T-cells by flow cytometry.Note: The staining procedure is summarized in Table 1.
-
a.Transfer 2.5 × 105 transduced and untransduced γδ[T2] cells into three separate 5 mL flow cytometry tubes.
-
b.Wash each tube with 2 mL PBS and centrifuge at 400 × g for 5 min.
-
c.Discard PBS and resuspend the cell pellet in 50 μL PBS.
-
d.Step 1: Add 5 μL anti-human γδ TCR-FITC antibody to one tube containing untransduced γδ[T2] cells (Ut tube 1).
-
e.Add 5 μL FITC-conjugated isotype control to the second tube containing untransduced γδ[T2] cells (Ut tube 2).
-
f.Add 5 μL anti-human γδ TCR-FITC antibody to all three tubes containing transduced γδ[T2] cells (Td tubes 1–3).
-
g.Place tubes on ice for 30 min.
-
h.Wash each tube with 2 mL FACS buffer and centrifuge at 400 × g for 5 min.
-
i.Discard PBS and resuspend cells in Ut tube 1, Ut tube 2 and Td tube 1 in 250 μL PBS. These tubes are now ready for analysis.
-
j.Add 50 μL PBS to Ut tube 3, Td tube 2 and Td tube 3.
-
k.Step 2: Add 3 μL biotinylated anti-human EGF antibody to Ut tube 3 and Td tube 3 and place on ice for 30 min in the dark.
-
l.Wash each tube with 2 mL PBS and centrifuge at 400 × g for 5 min.
-
m.Discard PBS and add fresh 200 μL PBS to Td tube 2. This tube is now ready for analysis.
-
n.Discard PBS and add 50 μL to Ut tube 3 and Td tube 3.
-
o.Step 3: Add 3 μL Streptavidin-PE to Ut tube 3, Td tube 2 and Td tube 3 and incubate on ice for 30 min in the dark.
-
p.Wash each tube with 2 mL PBS and centrifuge at 400 × g for 5 min and return to ice.
-
q.Discard PBS and resuspend cells in 250 μL PBS.
-
r.Analyze using flow cytometry.Note: A representative example of expression of pCAR-H/T in retrovirus transduced γδ[T2] cells is shown in Figure 6, together with efficiency of in vitro expansion of these cells and untransduced control cells.
-
a.
Figure 6.
Expression of the pCAR H/T parallel CAR in retrovirus-engineered γδ[T2] cells
The structure of pCAR H/T is shown schematically on the left. A representative example of flow cytometric analysis of γδ[T2] cells that express pCAR H/T are shown in the middle. Expansion of pCAR H/T γδ[T2] cells is shown on the right, expressed as fold increase in total cell number Note similar expansion efficiency to untrans(duced) γδ[T2] cells.
Table 1.
Staining of CAR expression in transduced γδ[2] and γδ[T2] cells
| Tube | Step 1 | Step 2 | Step 3 | Purpose |
|---|---|---|---|---|
| Ut1 | anti-human γδ TCR-FITC | Detect Ut γδ T-cells | ||
| Ut2 | Isotype FITC | Set baseline FITC signal to detect Ut γδ T-cells | ||
| Ut3 | Anti-EGF biotin | Streptavidin PE | Set baseline APC signal to detect transduced cells (highest of Ut3 vs Td2) | |
| Td1 | anti-human γδ TCR-FITC | Detect γδ T cells in transduced population. Should correlate with number identified in Td2 and Td3 tubes | ||
| Td2 | anti-human γδ TCR-FITC | Streptavidin PE | Set baseline APC signal to detect transduced cells (highest of Ut3 vs Td2) | |
| Td3 | anti-human γδ TCR-FITC | Anti-EGF biotin | Streptavidin PE | Detect transduced γδ T-cells |
Measuring in vitro tumor cytolytic activity and re-stimulation potential of γδ[T2] CAR T cells
Timing: 4 days minimum
This method quantifies tumor cell killing by pCAR γδ[T2] cells that target MUC1 and in which signaling is boosted by ErbB dimer co-expression, making comparison with untransduced γδ[T2] cells. The method can also be extended to assess tumor re-stimulation potential. This provides a measure of resistance of the CAR-engineered γδ T cells to exhaustion, induced by repeated antigen exposure (Vardhana et al., 2020).
Note: It is recommended to use tumor cell lines that have been passaged 20 times or less.
-
48.Day 1 – prepare tumor cells:
-
a.Seed tumor cells at 1 × 105 cells in 500 μL D10 medium per well in tissue culture treated 24 well plates.Note: Ensure that triplicate wells are plated for each T-cell population to be tested.
-
b.Incubate cells at 37°C & 5% CO2 for 24 h.Note: validate tumor cell surface expression of the target antigen(s) recognized by the CAR under study prior to co-culture experiments. In this example, firefly luciferase-expressing BxPC3 tumor cells have been used which co-express both MUC1 and multiple ErbB dimers (Whilding et al., 2017) and which have limited intrinsic sensitivity to killing by γδ T-cells.
-
a.
-
49.Day 2 – prepare T cells:
-
a.Count transduced and untransduced γδ[T2] cells. Re-suspend to a density of 2 × 105 cells per 0.5 mL TexMACSTM+ media.Note: This provides cells for a 2:1 E:T ratio. An excess should be prepared so that cells can be processed by serial 2-fold dilution.
-
b.Add 0.5 mL CAR or non-transduced γδ[T2] cells per well of tumor cells in triplicate.
-
c.Add 1 mL of TexMACSTM+ / D10 medium (equal parts mixture) to three wells to act as tumor alone control.
-
d.Incubate plate 37°C & 5% CO2.Note: No cytokine support is provided for T cells during the co-culture assay.
-
a.
-
50.
Day 3 (Optional) – collect supernatant for cytokine analysis as per step 31.
-
51.Day 4 – analyze cytotoxicity.
-
a.Add d-luciferin at 150 mg/mL immediately prior to luminescence reading.
-
b.Measure luminescence using appropriate apparatus and settings.
-
c.Calculate tumor cell viability using the following equation:
-
a.
-
52.
To assess tumor re-stimulation potential, repeat steps 48–51 twice per week until T cells can no longer be retrieved or less than 25% tumor cell killing occurs.
Note: Donor to donor variation is a significant issue in performing functional studies with primary human CAR T-cells. See troubleshooting section. An adequate number of biological replicates are required to ensure validity of results.
Note: Representative examples of tumor cell cytotoxicity and tumor cell re-stimulation on BxPC3 tumor cells are shown in Figure 7.
Figure 7.
BxPC3 tumor cell killing by pCAR H/T-engineered γδ[T2] cells
(A) Cytotoxicity assay in which untrans(duced) or pCAR-H/T transduced γδ[T2] cells were co-cultured at the indicated effector:target (E:T) ratio with ffLuc+ BxPC3 tumor cells for 48 h. Tumor cell viability was determined by luciferase assay (mean +/- SD, n=4).
(B) Replicate cytotoxicity assays conducted at a 1:1 E:T ratio for 48 h (mean +/- SD).
(C) Firefly luciferase BxPC3 tumor cells were co-cultivated with untrans(duced) or pCAR-H/T transduced γδ[T2] cells. Tumor cell viability was measured after 3–4 days. If >25% tumor cell killing occurred, γδ[T2] cells were re-stimulated by transfer to a new tumor cell monolayer. Total number of successful re-stimulation cycles is shown. Statistical analysis was performed using an unpaired Student’s t test.
Measuring in vivo antitumor activity of γδ[T2] CAR T cells
Timing: 5–16 weeks
This method quantifies in vivo anti-tumor activity of γδ[T2] cells in a challenging intraperitoneal (i.p.) xenograft model of pancreatic cancer, established using BxPC3 tumor cells. A representative example of such a study is shown in Figure 8. Tumor cells co-express red fluorescent protein (RFP) and firefly luciferase (ffLuc), achieved by retroviral transduction and flow sorting to purity (Whilding et al., 2017). Comparison is made with unmodified γδ[T2] cells that have negligible cytolytic activity against these tumor cells.
Note: It is recommended to use tumor cell lines that have been passaged 20 times or less.
Note: Group sizes should be determined prior to the experiment. In the example shown in Figure 8, a pilot study with 3 mice per group is shown. In definitive studies, a power calculation should be performed. One suitable online tool is found at http://www.biomath.info/power/ttest.htm (accessed February 1st, 2022). See troubleshooting step.
Note: To reduce bias, the experiment should be performed in a blinded fashion whereby one team member prepares γδ[T2] cells in coded tubes for administration by a blinded second team member.
Note: All in vivo experimentation must comply with local regulatory requirements. The experiment described here was performed using the authority of the U.K. Home Office project license number P23115EBF. The content of that license had been reviewed and approved by the King’s College London animal welfare and ethical review body (AWERB).
-
53.Day -2 – Isolate and activate PBMCs.
-
a.Perform steps 1–17/ 44–47 to expand sufficient numbers of pCAR-H/T transduced and untransduced γδ[T2] cells for each treatment group over a 2 week period.
-
a.
Note: CAR T-cell expansion data from previous experiments is required to calculate the number of γδ[T2] cells required for each group. In general, at least 10 million transduced cells can be generated from 1 million transduced γδ[T2] cells over 10 days when validated viral vector is used for the gene transfer step.
-
54.
6–10 week-old NSG mice are used for the experiment. In general, equal numbers of male and female mice should be used.
Note: Adequate time must be allowed for mice to acclimatize after delivery from a commercial supplier (generally up to 7 days).
-
55.Day 1 – Establish BxPC3 human pancreatic ductal adenocarcinoma xenograft in NSG mice.
-
a.Take baseline weight readings and check mice for signs of ill health prior to tumor injection.
-
b.Inject 1 × 105 ffLuc/RFP-expressing BxPC3 tumor cells suspended in 100 μL sterile PBS into all mice using the i.p. route.
-
c.Weigh mice twice weekly and check for signs of ill health daily.
-
a.
Note: Mice should be humanely killed using an appropriate procedure in the event of weight loss >15%, if total tumor flux exceeds 1 × 1010 photons per second (p/s) or if tumor growth impairs normal behavior or vital function. Abdominal distention, dyspnoea, neurological dysfunction, lameness, jaundice, piloerection, hunched posture, inability to groom, inactivity, and/ or inappetence are all additional humane end points and should prompt the humane killing of the affected animal.
-
56.Day 6 – Perform bioluminescence imaging (BLI) of mice.
-
a.Inject mice i.p. with 150 mg/kg D-luciferin.
-
b.Place mice into an anesthetic induction chamber (isoflurane 2.5%, with flow rate 2 L/min) until fully anesthetized.
-
c.Fifteen minutes after the D-luciferin injection, image the mice with an IVIS Spectrum Imaging platform (Perkin Elmer) with Living Image software, using the auto-exposure function.
-
d.Determine total flux (p/s) by creating a region of interest over each entire mouse.
-
e.After imaging, return the mice to their cages and monitor until they have fully recovered from anesthesia.
-
a.
-
57.Day 11 – Perform bioluminescence imaging of mice.
-
a.Mice that have total flux values greater than the enrollment threshold (usually >1 × 107 photons/second) are distributed into treatment groups with similar average tumor burden.
-
a.
-
58.
Day 12 – Inject the appropriate dose of transduced and untransduced γδ[T2] cells into each mouse using the i.p. route (10 × 106 cells in this example).
-
59.
Monitor the mice daily for signs of ill health.
-
60.
Weigh the animals twice weekly.
Note: Reduced weight, piloerection, ruffled coat, poor appetite, and reduced locomotion can all be signs of graft versus host disease (GvHD). Animals should be humanely killed if they show these signs without improvement for 48 h; if they lose >15% of body weight or if they develop other GvHD related signs such as diarrhea (>48 h).
-
61.
To monitor tumor status, weekly bioluminescence imaging is performed for the duration of the study.
-
62.
Mice with total flux (p/s) values reaching the humane endpoint threshold (1 × 1010 p/s) are humanely sacrificed by cervical dislocation or asphyxiation in CO2.
-
63.
Tumors are fixed in 10% formalin and paraffin embedded for future analysis.
CRITICAL: Prior to starting experiments, follow Perkin Elmer protocols to determine the D-luciferin kinetic curve for your model (https://resources.perkinelmer.com/lab-solutions/resources/docs/SOP_Determine_Luciferin_Kinetic_Curve.pdf, accessed January 2nd, 2022). Because bioluminescence emission is tissue dependent, optimal timings between injection of D-luciferin substrate and imaging should be determined prior to starting experiments. Serial imaging is performed every 5 min for a total of 40 min after injection of D-luciferin substrate. Total flux is plotted against time and this curve allows the determination of an imaging time at which maximum total flux is observed (most commonly at 10–20 min post injection).
Figure 8.
In vivo anti-tumor activity of pCAR H/T-engineered γδ[T2] cells
1 × 105 ffLuc+ BxPC3 tumor cells were injected i.p. in NSG mice. After 12 days, mice were treated with 10 × 106 untrans(duced) or pCAR-H/T transduced γδ[T2] cells, making comparison with PBS. Serial BLI emission is shown (mean +/- SD, n=3). Statistical analysis was performed using an unpaired Student’s t test.
Expected outcomes
It is anticipated that γδ[T2] cells will have enhanced viability and yield, accompanied by increased intrinsic anti-tumor cytolytic activity. Activation of γδ[T2] cells is accompanied by increased cytokine production compared to counterparts expanded in IL-2 alone. In vivo anti-tumor activity of these cells is also expected to be increased and in a manner that can be targeted using a chimeric antigen receptor (Beatson et al., 2021).
Quantification and statistical analysis
Statistical significance is determined using GraphPad Prism 9 software. Statistical comparison between two groups is undertaken using an unpaired Student t-test, assuming normally distributed data. Comparison of three or more groups is performed using one-way or two-way ANOVA with multiple comparisons, when one or two independent variables were present respectively.
Limitations
Donor variation, in terms of number, purity and phenotype of ex vivo expanded γδ T cells is substantial and there is a need to identify attributes/ biomarkers of the starting material that are predictive of robust expansion of cells with the greatest intrinsic anti-tumor activity.
In vivo analysis of human CAR T cell functionality using immunocompromised mice is limited by many factors. Since NSG mice do not have human hematopoietic cells or an intact immune system, it is difficult to predict how CAR T cells might overcome immunosuppressive cellular factors, particularly in the case of solid malignancies. Moreover, in an NSG model, CAR T cells may not be exposed to the same immune suppressive checkpoint molecules, cytokines, and metabolic stresses which may be present in a naturally occurring tumor.
Using healthy donors as a source of PMBCs often results in successful transduction and expansion of CAR γδ[T2] cells. However, in a clinical setting when using autologous patient T cells, prior exposure to treatments such as chemotherapy and radiotherapy may compromise the quality and yield of CAR γδ[T2] cells.
Troubleshooting
Problem 1
Low yields of ex vivo expanded γδ T cells (steps 1–17).
Potential solution 1
It is appropriate to check starting material to quantify the number of γδ T cells that are present. Upon culture on immobilized anti-γδ TCR antibody, clustering and enlargement of T cell size are generally visible microscopically. Similar attributes are observed in ZOL-activated cultures after a few days. We have found that freshly isolated PBMC from healthy donors are the most reliable source of these cells. Feeder cells that provide mitogenic and/or co-stimulatory signals to the cells may also be used to enhance cell yield (Xiao et al., 2018).
Problem 2
Lack of distinctive phenotype associated with γδ[T2] cells (step 17). γδ[T2] cells typically have a highly distinctive phenotype with high expression of CD103, CXCR3, E-selectin binding activity and high levels of cutaneous leukocyte antigen (Beatson et al., 2021).
Potential solution 2
The quality of the TGFβ1 used to supplement the culture should be checked, if necessary, using a functional assay (Abe et al., 1994). It is also important to closely follow manufacturer’s instructions, especially the addition of hydrochloric acid, when reconstituting this cytokine.
Problem 3
Tumor engraftment may fail in NSG mice following missed i.p. injection. Similarly, false negative BLI emission values may be obtained for the same reason (see steps 55–63).
Potential solution 3
Very few tumor cell lines will have 100% engraftment rates. Tumor cells left in PBS for extended periods of time before injection will greatly reduce engraftment rates. It may be useful to inject tumor in 3–4 additional mice to ensure that group sizes are sufficient for meaningful analysis. Tumor growth in PBS-treated mice tends to be most homogeneous meaning that a smaller group size may be tolerated than is the case for T cell-treated mice. If all animals engraft tumor successfully, it is recommended to add additional mice to the key T cell treatment groups. Suspected failed i.p. injection of D-luciferin is generally readily identifiable because of the trend of BLI emission in affected mice. If repeat luciferin injection yields a positive reading, the spurious negative value should be omitted from data analysis.
Problem 4
Retroviral transduction of γδ[T2] cells may be of sub-optimal efficiency (steps 44–47).
Potential solution 4
High quality plasmids are required for efficient transfection of HEK293T cells (see Larcombe-Young et al., currently under review as a STAR Protocol). HEK293T cells should be of low passage number when transfected. γδ[T2] cells should be activated on the day of gene transfer, indicated by formation of clusters/ clumps of enlarged T cells. If problems persist, it may be appropriate to titrate viral vector in order to ensure that batches contain a high titer of virus.
Problem 5
Undertaking functional in vivo studies using CAR-engineered γδ[T2] cells can be unpredictable due to donor-to-donor variability (see steps 53–63). T cell fitness, state of differentiation and yield can vary significantly between donors.
Potential solution 5
The primary solution to this issue is to perform each experiment with multiple biological replicates.
Problem 6
Cytotoxicity assays are conveniently performed using luciferase assays as described in steps 48–51 of this protocol. However, this method requires the stable expression of firefly luciferase in the tumor cells.
Potential solution 6
If luciferase-expressing tumor cells are unavailable, MTT assays can be performed to quantify cytotoxicity of T cells against adherent tumor cell monolayers. This measures the reduction of tetrazolium in a manner that is dependent on intact mitochondrial function. An example of the use of this assay is provided in steps 23–27.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Dr John Maher (john.maher@kcl.ac.uk).
Materials availability
Constructs and other reagents generated or described in this study will be made available from the lead contact for academic/noncommercial research purposes on request. Commercial use of the constructs generated, or derivatives, would be subject to a licensing agreement as intellectual property rights are in place.
Acknowledgments
This study was funded by Bloodwise (Programme 13007), Cancer Research UK (CRUK)/ Bayer (C11499/A19444), the Medical Research Council (DPFS MR/ M024733/1 and MR/R000026/1), a Medical Research Council Confidence in Concepts award, a CRUK Multidisciplinary Project Award (C48390/A21153), the British Lung Foundation (MESOUK18-2), the Experimental Cancer Medicine Centre at King’s College London, the King’s Health Partners/ King’s College London Cancer Research UK Cancer Centre, Leucid Bio and by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust, and King’s College London (grant number IS-BRC-1215-20006). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. We thank many colleagues for generously providing cell lines.
Author contributions
A.P-P., R.B., D.D., C.H., and L.W. conducted experimental work. A.P-P., R.B., D.D., C.H., L.W., and J.M. designed and optimized the approach and interpreted the data. R.B. and J.M. wrote the first draft of the manuscript. All authors reviewed and edited the manuscript.
Declaration of interests
J.M. is scientific founder, chief scientific officer, and shareholder in Leucid Bio which is a spin-out company focused on development of cellular therapeutic agents. J.M., A.P-P., and R.B. are named inventors on granted patents pertaining to this work. L.W. and D.M.D. were both formerly consultants to Leucid Bio. J.M. and L.W. are named inventors on a granted patent submitted in relation to this work while D.D., C.H., L.W., and J.M. are named inventors on submitted patents that relate to pCAR technology. D.M.D. is a shareholder in Leucid Bio. J.C.P. declares no competing financial interests.
Data and code availability
The datasets supporting this protocol, and constructs described or used in Figures, have not been deposited in a public repository but are available from the corresponding author upon request.
References
- Abe M., Harpel J.G., Metz C.N., Nunes I., Loskutoff D.J., Rifkin D.B. An assay for transforming growth factor-beta using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 1994;216:276–284. doi: 10.1006/abio.1994.1042. [DOI] [PubMed] [Google Scholar]
- Beatson R.E., Parente-Pereira A.C., Halim L., Cozzetto D., Hull C., Whilding L.M., Martinez O., Taylor C.A., Obajdin J., Luu Hoang K.N., et al. TGF-beta1 potentiates Vgamma9Vdelta2 T cell adoptive immunotherapy of cancer. Cell Rep. Med. 2021;2:100473. doi: 10.1016/j.xcrm.2021.100473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieli F., Poccia F., Lipp M., Sireci G., Caccamo N., Di Sano C., Salerno A. Differentiation of effector/memory Vdelta2 T cells and migratory routes in lymph nodes or inflammatory sites. J. Exp. Med. 2003;198:391–397. doi: 10.1084/jem.20030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D., et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabelitz D., Serrano R., Kouakanou L., Peters C., Kalyan S. Cancer immunotherapy with gammadelta T cells: many paths ahead of us. Cell Mol Immunol. 2020;17:925–939. doi: 10.1038/s41423-020-0504-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muliaditan T., Halim L., Whilding L.M., Draper B., Achkova D.Y., Kausar F., Glover M., Bechman N., Arulappu A., Sanchez J., et al. Synergistic T cell signaling by 41BB and CD28 is optimally achieved by membrane proximal positioning within parallel chimeric antigen receptors. Cell Rep. Med. 2021;2:100457. doi: 10.1016/j.xcrm.2021.100457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhana S.A., Hwee M.A., Berisa M., Wells D.K., Yost K.E., King B., Smith M., Herrera P.S., Chang H.Y., Satpathy A.T., et al. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol. 2020 doi: 10.1038/s41590-020-0725-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whilding L.M., Parente-Pereira A.C., Zabinski T., Davies D.M., Petrovic R.M.G., Kao Y.V., Saxena S.A., Romain A., Costa-Guerra J.A., Violette S., et al. Targeting of aberrant alphavbeta6 integrin expression in solid tumors using chimeric antigen receptor-engineered T cells. Mol. Ther. 2017;25:259–273. doi: 10.1016/j.ymthe.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Chen C., Li Z., Zhu S., Tay J.C., Zhang X., Zha S., Zeng J., Tan W.K., Liu X., et al. Large-scale expansion of Vgamma9Vdelta2 T cells with engineered K562 feeder cells in G-Rex vessels and their use as chimeric antigen receptor-modified effector cells. Cytotherapy. 2018;20:420–435. doi: 10.1016/j.jcyt.2017.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting this protocol, and constructs described or used in Figures, have not been deposited in a public repository but are available from the corresponding author upon request.