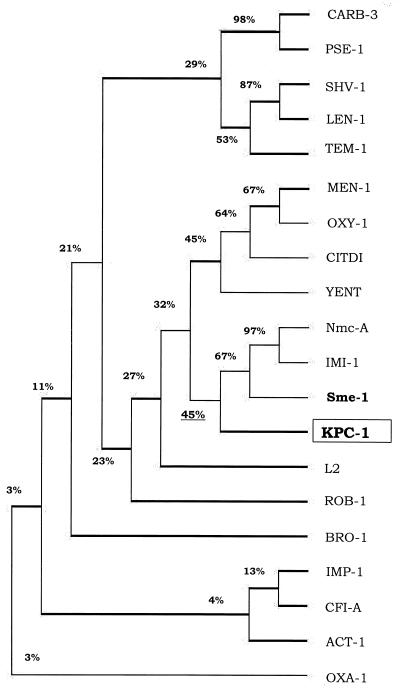
FIG. 6.
Dendrogram showing similarity of 20 β-lactamases. The dendrogram was constructed by using DNASIS for Window's multiple alignment option (Higgins-Sharp). Sixteen of the β-lactamases are class A enzymes, CARB-3 from P. aeruginosa (27), PSE-1 from P. aeruginosa (24), SHV-1 from E. coli (39), LEN-1 from K. pneumoniae (2), TEM-1 from E. coli (65), MEN-1 from E. coli (6), OXY-1 from Klebsiella oxytoca (19), CITDI from C. diversus (50), YENT from Y. enterocolitica (61), Nmc-A from E. cloacae (42), IMI-1 from E. cloacae (57), Sme-1 from S. marcescens (41), L2 from Stenotrophomonas maltophilia (71), ROB -1 from Haemophilus influenzae (31), and BRO-1 from Moraxella catarrhalis (D. Beaulieu, L. Piche, T. R. Parr, Jr., K. Roeger-Lawry, P. Rosteck, and P. H. Roy, β-lactamase BRO-1 precursor [penicillinase], gi:2497581, Gen Bank, 1996); IMP-1 from S. marcescens (48) and Cfi-A from Bacteroides fragilis (69) were included as representatives of class B (metallo-β-lactamases); ACT-1 from K. pneumoniae (7) represents class C, AmpC β-lactamases; and OXA-1 from E. coli (49) represents class D enzymes.