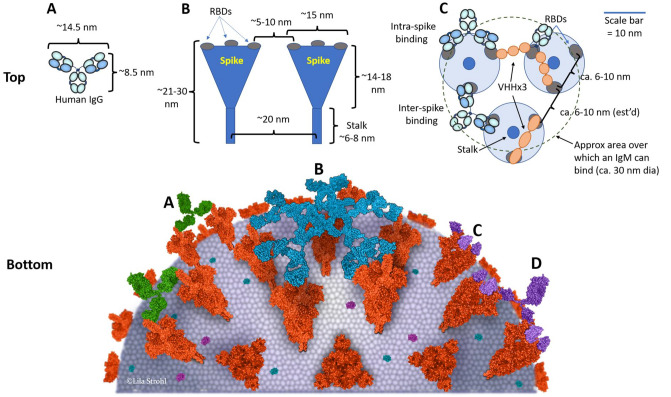
Fig. 11.
A model of the surface of a SARS-CoV-2 virion showing sizes and distances of spike and antibodies. Top: A The size and shape of a human IgG1 [480]. B The size, shape, and dimensions of SARS-CoV-2 spike proteins [361], informed also by SARS-CoV-1 spike protein anatomy [485, 486]. There are approximately 100 spikes per 90 nm diameter virion [361]. C A model showing how the sizes and spacings allow for potential inter-spike cross-linking by both IgGs, as recently observed by Hastie et al. [290], as well as potential for crosslinking by triple-VHH-Fc fusions. This model also shows the approximate coverage of an IgM, which has a diameter of about 30 nm [337]. Bottom: Model of SARS-CoV-2 targeted by antibodies. SARS-CoV-2 spike proteins with one open RBD or all closed RBDs based on PDB ID 6VYB and 6VXX, respectively. A IgGs (PDB ID 1IGT) crosslinking two spikes; B an IgM (based on PDB ID 2RCJ) binding to multiple spikes similar to a hand palming a basketball; C a trimeric VHH-Fc fusion protein (based on PDB IDs 6ZXN and 1IGT as per Fig. 2) cross-linking spikes; and D individual VHHs (PDB ID 6ZXN) binding to RBDs with no cross-linking ability. For the Bottom drawing, the PDB program [201, 202] was used to generate the structures. Approx approximate, dia diameter, IgG immunoglobulin G, IgM immunoglobulin M, nm nanometers, PDB ID Protein Data Bank Identifier, RBD receptor binding domain, SARS-CoV-2 severe acute respiratory syndrome coronavirus-2, VHH single domain antibody, VHH-Fc single domain antibody fused to an IgG-Fc domain