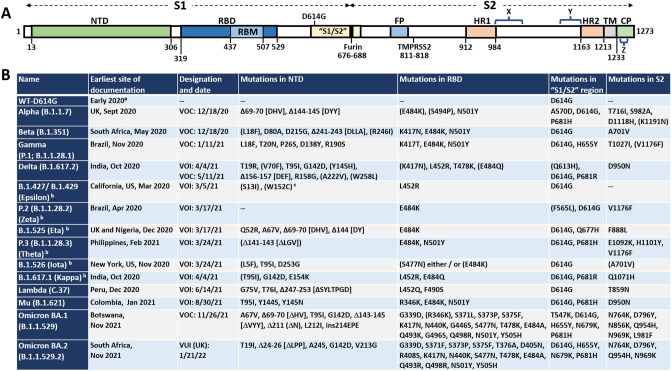
Fig. 2.
Locations of mutations in SARS-CoV-2 spike protein. A Linear representation of SARS-CoV-2 spike protein showing some of the key subunits involved with mutations as well as antibodies, including the N-terminal domain (NTD), the receptor binding domain (RBD), receptor binding motif (RBM), S1/S2 region around the furin protease cleavage site, the S2 domain, and the transmembrane (TM) region at the C-terminus of S2. Notes: X, residues 980-1006 are an epitope for S2-targeting neutralizing antibody 3A3; Y, residues 1140–1164 in stem-helix region targeted by antibody CC40.8; Z, residues 1229–1243 in stem-helix region targeted by neutralizing antibody 28D9. B A complete list of all the World Health Organization (WHO) designated Variants of Concern (VOC) and the Variants of Interest (VOI) as well as the “original variant” D614G, and the mutations each variant carries in each of the major domains. All amino acids are noted by their single letter designation. NC not categorized. Deletions are noted by Δ followed by the deleted amino acids. aD614G was found in many sequences very soon after sequencing efforts began in early 2020; b the VOIs Epsilon, Theta, Eta, Kappa, Iota, and Zeta, were declassified as VOIs, so those names are provided parenthetically; cmutant positions in parentheses (e.g., (S13I)) indicate mutations that are only sometimes associated with the variant listed. Data are from the WHO [31] and US Centers for Disease Control (US CDC) [32]. Data for Omicron are from the US CDC [33]