An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.
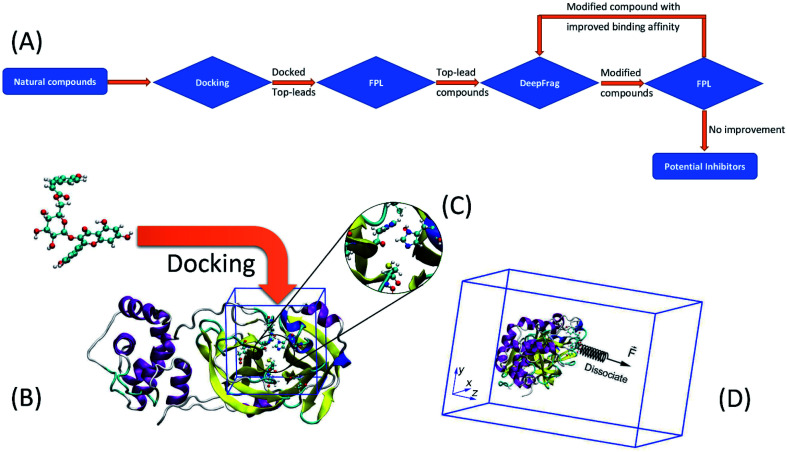
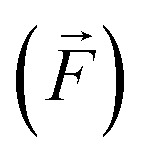 during FPL simulations.
during FPL simulations. 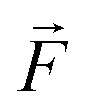 was put on the ligand center of mass in order to force the ligand to mobilize out of the protease binding cavity.
was put on the ligand center of mass in order to force the ligand to mobilize out of the protease binding cavity.