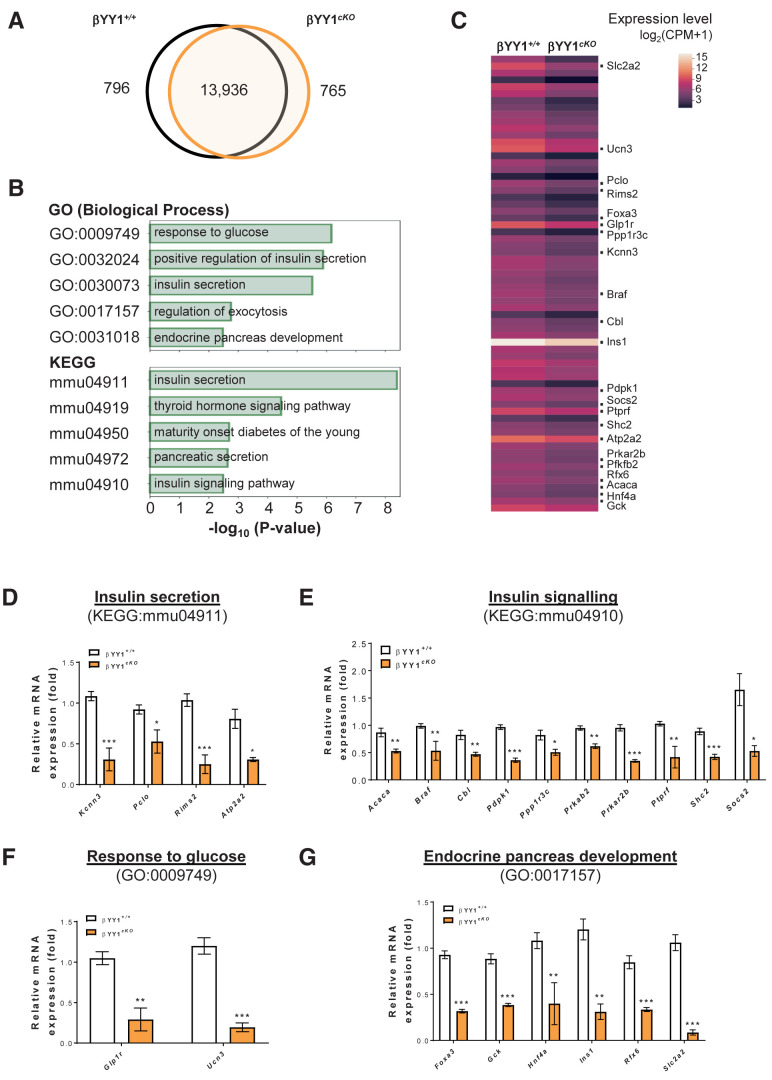
Figure 4.
Genome-wide RNA-seq profiling reveals impaired glucose response and INS signaling after β-cell loss of Yy1. Tam was administered to adult Pdx1CreERT/+;Yy1fl/fl mice within 2 weeks of the start of the experiment, and analyses were performed at 2 weeks after Tam treatment. (A) Venn diagram illustrating transcripts that are significantly differentially expressed (log2fc ≥1 or log2fc ≤−1; false discovery rate–adjusted P value ≤0.05) in islets isolated from β-cell Yy1-deficient mice (i.e., cKO mice) compared with that of the Yy1fl/fl (control) mice. A total of 1,561 transcripts were altered in abundance in vivo, including 796 downregulated and 765 upregulated genes in cKO compared with control islets. (B) The top 5 most significant pathways by GO in terms of biological processes or KEGG pathways determined by downregulated genes expressed by the pancreatic islets of cKO compared with that of control mice. (C) Heat map of selected differentially expressed genes as determined by GO and KEGG functional clustering from (B). The red scale bar denotes the log2 expression level of genes in each group. (D–G) Quantitative RT-qPCR of the highlighted genes in terms of (D) INS secretion, (E) INS signaling, (F) response to glucose, and (G) endocrine pancreas development with reference to (C). Gene expression levels in cKO islets were compared with that of the control islets. Data are presented as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; n = 6 mice per group.