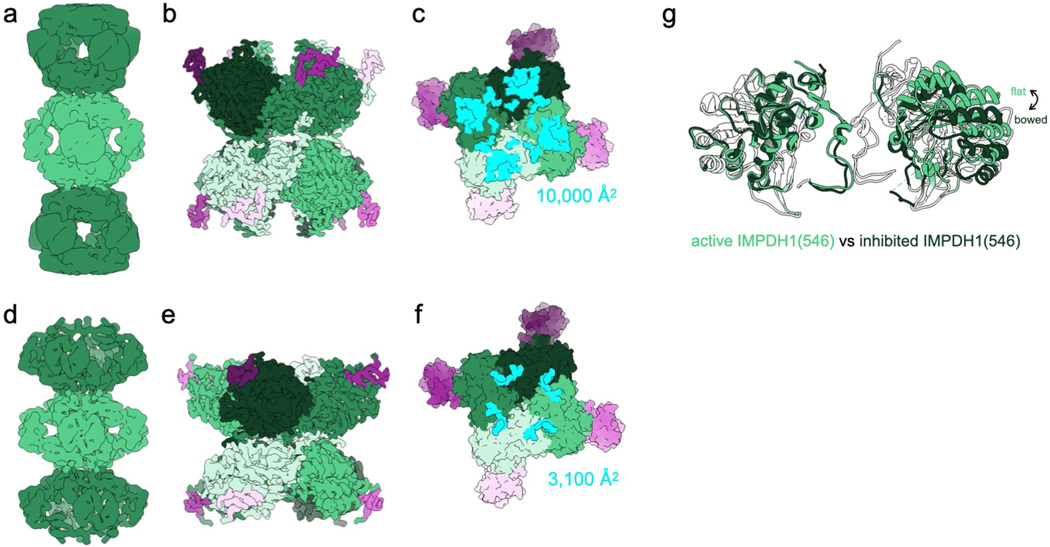
Extended Data Fig. 8 |. IMPDH1 retinal variant (546) is similar to canonical IMPDH1.
a-c, Active IMPDH1(546) filament bound to ATP, IMP, nAD+. a, Low-pass filtered cryo-EM reconstruction b, Interface-focused cryo-EM reconstruction. 8 monomers are colored by catalytic domain (green) and regulatory domain (pink). c, View of the top of an octamer from inside the filament. The surface area buried by the octamer interface is in aqua with the indicated total buried surface area. (Surface representation of the atomic model at the assembly interface, with buried residues in cyan). d-f, Inhibited IMPDH1(546) filament bound to GTP, ATP, IMP, nAD+. d, Low-pass filtered cryo-EM reconstruction e, Interface-focused cryo-EM reconstruction. 8 monomers are colored by catalytic domain (green) and regulatory domain (pink). f, View of the top of an octamer from inside the filament. The surface area buried by the octamer interface is in aqua with the indicated total buried surface area. (Surface representation of the atomic model at the assembly interface, with buried residues in cyan).