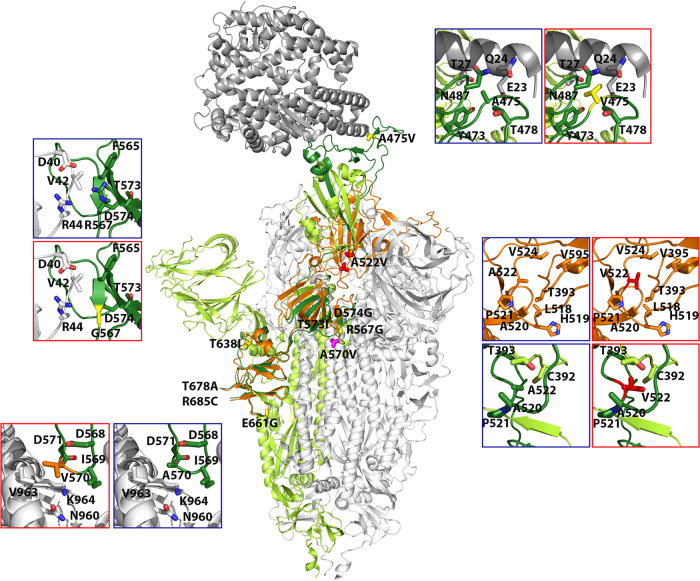
FIG 6.
Location of amino acid substitutions in the three-dimensional structure of spike (S) protein. The central structure is a cartoon representation of S trimer (PDB code 7A94) with the reference monomer colored in green and dark green, the latter marking the regions covered by amplicons A5 to A6 (indicated in Fig. 1). The remaining monomers of the S trimer are shown in gray. The reference monomer contains the RBD domain in the “erect” position. A superimposition of this domain in the “open” conformation is also shown in orange. Substitutions are labeled, and amino acids are shown as sticks in different colors, according to associated disease category: exitus in red; moderate in magenta, and mild in yellow. Insets highlight the interactions of some substituted positions with neighboring residues within a 5-Å radius. Except for position 522, two insets are shown per mutated position, indicating the original and mutated residues, squared in blue and red, respectively. For position 522, four insets are shown; the top two indicate the interactions of this residue in the open conformation of RBD, and the bottom two indicate those in the erect conformation. The substitutions and their frequency in the mutant spectrum, acceptability, functional score, and possible structural or functional effects are listed in Table 3.