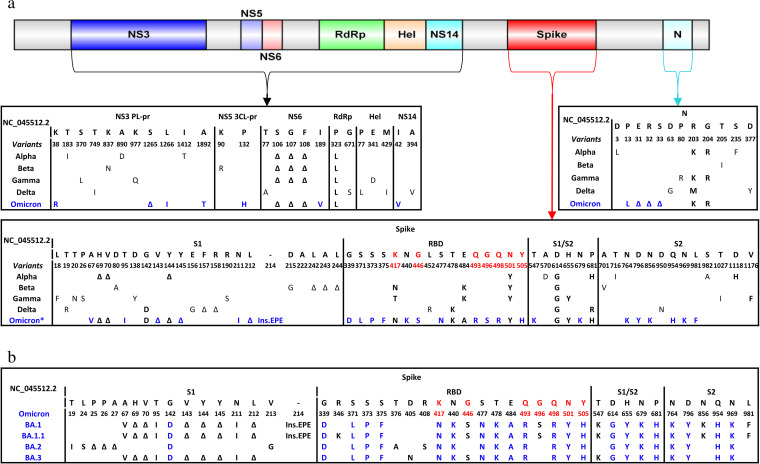
FIG 1.
(a) Schematic illustration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome and its variants of concern along with their relevant mutations that characterize each protein of interest. Mutations were defined as amino acid substitutions/deletions/insertions that occurred in 75% of sequences using the NC_045512.2 as the reference sequence. Residues in red are those that directly interact with ACE2. Mutations in bold black refer to the shared mutations by at least two variants of concern, while those in bold blue refer to the unique Omicron mutations. *Mutations at residues 417, 440, and 446 in the receptor-binding domain (RBD) spike represent original Omicron consensus according to earlier prevalence of greater than 75%, while currently, the new prevalence values are 51.3, 53.9, and 54,9%, respectively. (b) Spike mutations underlying the currently circulating Omicron sublineages BA.1, BA.1.1, BA.2, and BA.3. Blue color refers to those mutations shared by the four sublineages.