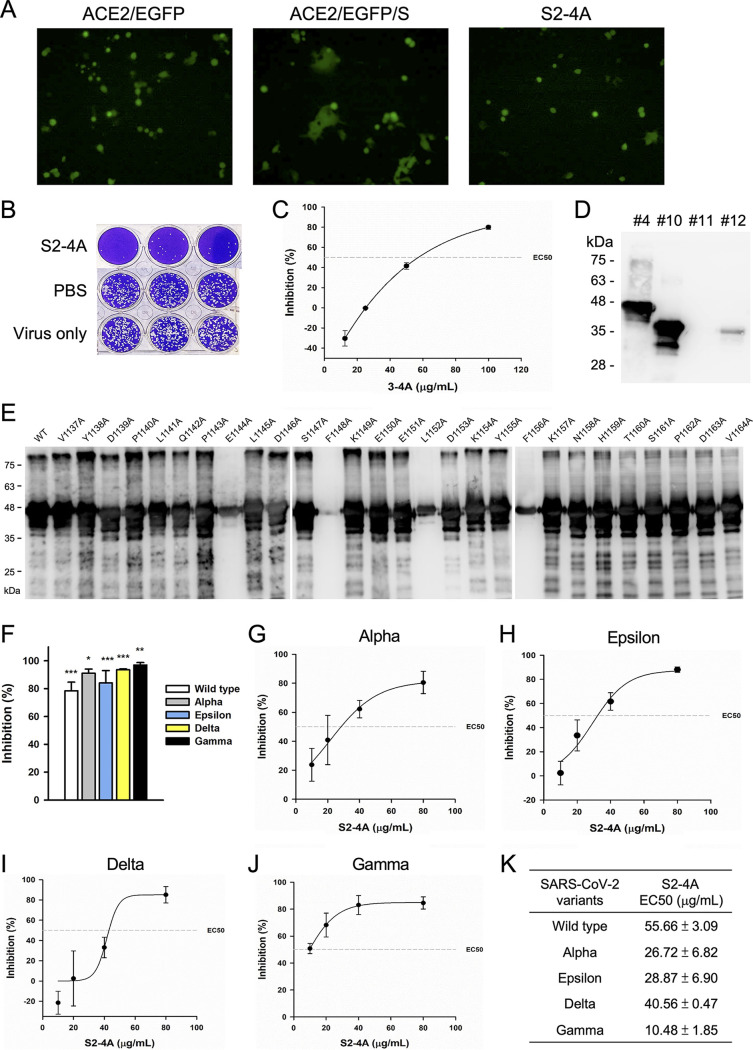
FIG 7.
The virus neutralizing activity and antigenic specificity of MAb S2-4A. (A) The S protein-mediated syncytium formation assay was performed to analyze the neutralizing activity of MAb S2-4A. The formation of the syncytia by coculturing the EGFP-transfected ACE2-293T cells with the S protein-transfected 293T cells was markedly inhibited in the presence of S2-4A (100 μg/mL). (B) The MAb S2-4A (100 μg/mL) was incubated with the SARS-CoV-2 wild-type strain. and the plaque reduction assay was applied to evaluate the neutralizing activity. Data are three biological replicates (n = 3). (C) The EC50 of MAb S2-4A against SARS-CoV-2 was determined by using a series of MAb S2-4A (100, 50, 25, 12, 5 μg/mL) in the plaque reduction assay. (D) The binding of MAb S2-4A to S2 truncation fragments #4, #10, #11, and #12 was determined by WB analysis. (E) The sfGFP-S(1042-1167) mutants with a series of individual alanine substitution at residues 1137–1164 were analyzed on SDS-PAGE and then subjected to WB analysis with MAb S2-4A to identify the critical antigenic determinants. (F) The S2-4A (100 μg/mL) was preincubated with the SARS-CoV-2 wild-type strain (white bar), Alpha (gray bar), Epsilon (blue bars), Delta (yellow bars), and Gamma (black bars) variants and then subjected to the plaque reduction assay. The inhibition of the plaque formation by MAb S2-4A was calculated by comparing to the experimental results in the absence of antibodies. Data are presented as means ± SD of three biological replicates (n = 3). One-way ANOVA was used for statistical analysis. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (G–J) For determination of the EC50 values of S2-4A against SARS-CoV-2 Alpha, Epsilon, Delta, and Gamma variants, a series of diluted MAb S2-4A solutions (80, 40, 20, and 10 μg/mL) were utilized in the plaque reduction assay. Data are presented as means ± SD of three biological replicates (n =3), and further graphed by linear regression. (K) The EC50 values of S2-4A against different SARS-CoV-2 variants.