Figure 4. ATP8B1-CDC50A is autoinhibited by both its N- and C-terminal tails and the presence of lipids is required for its activity.
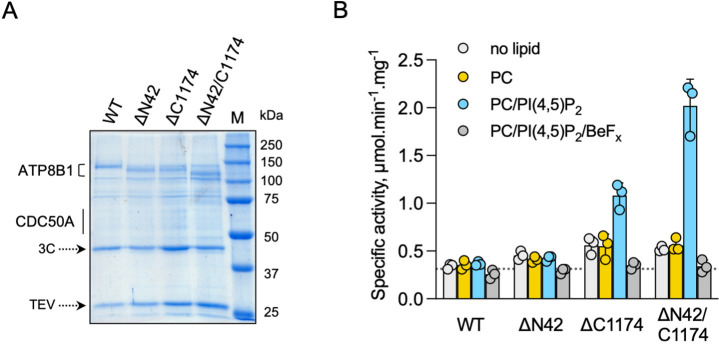
(A) Removal of N- and/or C-terminal extensions of ATP8B1 upon on-column cleavage of streptavidin-bound ATP8B1-CDC50A with both TEV and 3 C proteases assessed by Coomassie blue stained SDS-PAGE. ΔN42 lacks residues 1–42 of ATP8B1 whereas ΔC1174 lacks residues 1175–1251 and ΔN42/C1174 lacks both. M, molecular weight marker. Streptavidin-purified wild-type (WT) and truncated mutants were used for subsequent ATPase assays. (B) ATPase activity of wild-type (WT), N-terminally truncated (ΔN42), C-terminally truncated (ΔC1174) and both N- and C-terminally truncated (ΔN42/C1174) ATP8B1 ( ~ 5 µg ml–1 protein) in complex with CDC50A determined in DDM at 30 °C. The assay medium contained 1 mM MgATP, 0.5 mg ml–1 DDM, and 0.01 mg ml–1 CHS. PC and PI(4,5)P2 were added at 0.1 mg ml–1 (132 µM) and 0.025 mg ml–1 (23 µM), respectively, resulting in a DDM final concentration of 1.25 mg ml–1. The PC/PI(4,5)P2 ratio is therefore 5.8 (mol/mol) Data are a mean ± s.d. of three technical replicate experiments (purification #1, see Materials and methods). The dotted line represents background NADH oxidation due to photobleaching, measured in the absence of purified protein and lipids. Source files related to (B) are available in Figure 4—source data 1.
Figure 4—figure supplement 1. Sequence alignment of selected P4-ATPases.
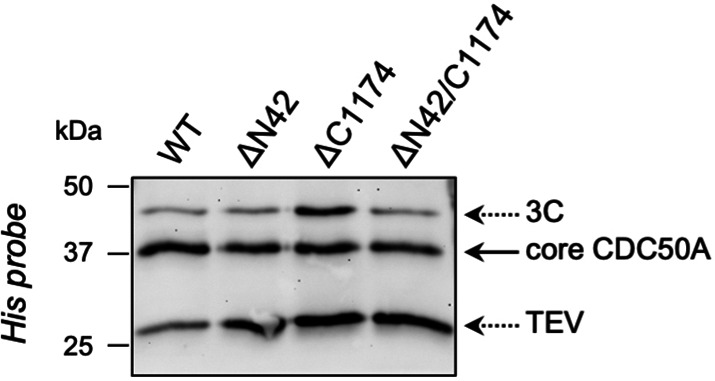
Figure 4—figure supplement 2. The amount of CDC50A which co-elutes with ATP8B1 upon on-column cleavage with TEV and 3 C proteases is similar for wild-type ATP8B1 (WT) and the 3 C protease cleavage site insertion mutants (ΔN42, ΔC1174, ΔN42/C1174).